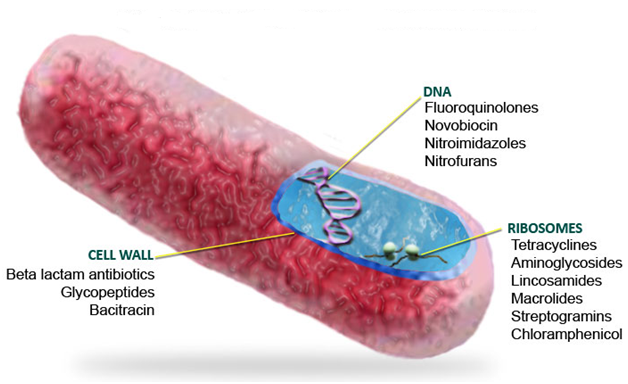
The antibiotics described above including those not described in this work are used to treat infections caused by disease causing microorganisms (bacteria). Majority of them exert a highly selective toxic action upon their target microbial cells but have little or no toxicity towards mammalian cells. These antibiotics can therefore be administered at concentrations sufficient enough to kill or inhibit the growth of infecting organisms without damaging mammalian cells. The ways by which these antibiotics exert their antibacterial activities on microbes in vivo without necessarily harming the host (patient) taking the drug is called the “Mechanism of Action of Antibiotics”.
It reveals and explains the rationale behind the selective toxicity of antibiotics and how they stop the venomous effects of bacteria. Antimicrobial agents (in particular antibiotics) show a wide variety of mechanisms of action against pathogenic microorganisms either in vivo or in vitro; and these shall be discussed in this section. The mechanism of action elaborated here is strictly for antibacterial agents (i.e. drugs that target pathogenic bacteria).
INHIBITION OF MICROBIAL CELL WALL SYNTHESIS: Peptidoglycan is a vital component of the cell wall of virtually all bacteria. But it is more pronounced in Gram positive bacteria than in Gram negative bacteria. The peptidoglycan is responsible for maintaining the shape and mechanical strength of the bacterial cell wall. If it is damaged in anyway, or its synthesis is inhibited (e.g. by antibiotics), then the shape of the bacterial cells becomes distorted and they will eventually burst (lyse) due to the high internal osmotic pressure following the influx of fluids or substances like drugs from the outside into the bacterial cell. Mammalian cell is devoid of peptidoglycan, thus antibiotics which inhibit microbial cell wall synthesis show outstanding selective toxicity. Examples of antibiotics that inhibit bacterial cell wall synthesis include: Penicillins, Bacitracin, Glycopeptides (e.g. Vancomycin), and Cephalosporins.
These antibiotics stop the cross-linking of N-acetyl-glucosamine (NAG) and N-acetyl-muramic acid (NAM) via a reaction known as transpeptidation reaction; and this cross-linking is supposed to lead to the formation of peptidoglycan, an important component of bacterial cell wall. Bacterial resistance to beta-lactam antibiotics (e.g. penicillins and cephalosporins) is one of the mechanisms used by pathogenic bacteria (both Gram-positive and Gram-negative organisms) to evade the antimicrobial onslaught of beta-lactam drugs; and this is because these antibiotics (i.e. beta-lactams) are the most widely used drugs in clinical medicine. However, the growing level of bacterial resistance towards these potent antibiotics is fast slowing the efficacy and clinical applications of these agents. Beta-lactam antibiotics as earlier explained are a class of drugs that inhibit the synthesis of cell wall in pathogenic bacteria.
The antibacterial activity of beta-lactams is exhibited in vivo when the drugs bind to specific receptors on the cell membrane or cell wall of the target bacterium especially the penicillin-binding-proteins (PBPs). Binding of beta-lactam antibiotics to the PBPs of pathogenic bacteria prevent the cross-linking of NAM and NAG. NAG and NAM are both vital for the synthesis of peptidoglycan, an important component of bacterial cell wall. The synthesis of peptidoglycan is inhibited once the cross-linking of NAM and NAG is hindered via the antibacterial action of beta-lactam antibiotics. And this renders the bacterial cell wall porous to external harmful substances such as drugs, water and other chemicals. The bacterial cell eventually dies following rupture or lysis of the cell. Nevertheless, pathogenic bacteria produce antibiotic-hydrolyzing enzymes such as beta-lactamases which render beta-lactam drugs inefficacious in vivo.
Beta-lactam antibiotics are unique because they have a ring known as the beta-lactam ring, and which is the target of the beta-lactamases produced by pathogenic bacteria. The beta-lactamase enzyme cleaves the carbon-nitrogen (C-N) bond of the beta-lactam ring of beta-lactams; and this eventually leads to the production of a compound with a lesser antibacterial activity. In the case of penicillin for example, cleavage of the beta-lactam ring of the penicillins leads to the formation of penicilloic acid which is devoid of any antibacterial activity. Pathogenic bacteria that express beta-lactamases as well as other expanded spectrum enzymes such as extended spectrum beta-lactamases (ESBLs) amongst others render beta-lactam antibiotics in vivo because of the antibiotic-hydrolyzing enzymes which they produce.
INHIBITION OF DNA SYNTHESIS FUNCTION: Deoxyribonucleic acid (DNA) is a vital component of the chromosome of bacterial cells as they are known to direct the activity of the cells. Antibiotics that inhibit the synthesis of DNA in bacterial cell work by interfering with the transcription stages which are very vital to the final and complete formation of bacterial DNA. These antibiotics block the DNA gyrase enzyme (Topoisomerase II) which is vital for the complete synthesis of DNA in bacteria. Examples of antibiotics that inhibit bacterial DNA synthesis function include: sulfonamides, trimethoprim, rifampicin, quinolones, and fluoroquinolones.
DESTRUCTION OF MICROBIAL CELL MEMBRANE: The integrity of the cytoplasmic membrane in bacterial cell is very important for the normal functioning of all cells. Bacterial cell membranes do not contain sterols. This differentiates them from fungal and mammalian cells which do contain sterols, thus giving such antibiotics a highly selective toxicity on bacteria. Antibiotics that destroy the cytoplasmic membrane of bacterial cells cause irreversible leakage of cytoplasmic components by disturbing the integrity of the membrane. This can also impair other metabolic functions associated with the membrane. Examples of antibiotics that destroy microbial cell membrane include: polymyxins, polyenes.
INHIBITION OF PROTEIN SYNTHESIS: Bacterial ribosomes are smaller than their mammalian counterparts. The ribosomes in bacterial cells are vital for the synthesis of proteins. Antibiotics that inhibit the synthesis of protein in bacterial cells act by binding to a receptor on either the 30S or 50S subunit ribosomes. This action prevents the complete reaction of translocation, thereby inhibiting the synthesis of protein in bacterial cells. Antibiotics that are protein synthesis inhibitors include: Tetracyclines, Chloramphenicol, Macrolides, and Streptomycin.
INHIBITION OF METABOLIC PATHWAY: The inhibition of a metabolic pathway in microbes (e.g. a pathway responsible for synthesizing key metabolites such as folic acid) is carried out by a group of antibiotics known as anti-metabolites. These antibiotics inhibit the production of key metabolites in their target organisms. Antibiotics that are anti-metabolites include: sulphonamides, trimethoprim and pyrimethamine. Anti-metabolites block the key steps in folate synthesis. Folate is an important cofactor in the biosynthesis of nucleotides which are the building blocks of DAN and RNA in microorganisms. Sulphonamides are structural analogues of Para-aminobenzoic acid (PABA). PABA is a key component in the synthesis of folic acid in bacteria. They competitively block the conversion of pteridine and PABA to dihydrofolic acid because the anti-metabolite, sulphonamides has a greater affinity for the enzyme (dihydropterate synthetase) that performs the conversion than does PABA. Bacteria and protozoa unlike most mammalian or animal cells (which obtain their own folic acid from their diet) synthesize their own folic acid. Sulphonamides competitively inhibit the incorporation of PABA into dihydropteroic acid (folic acid), and once this is done, folic acid will no longer be available for the synthesis of purines and pyrimidines which are both required for the coupling of bacteria DNA and RNA. This inhibits the synthesis of nucleic acid in the organism.
References
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Bisht R., Katiyar A., Singh R and Mittal P (2009). Antibiotic Resistance – A Global Issue of Concern. Asian Journal of Pharmaceutical and Clinical Research, 2 (2):34-39.
Courvalin P, Leclercq R and Rice L.B (2010). Antibiogram. ESKA Publishing, ASM Press, Canada.
Denyer S.P., Hodges N.A and Gorman S.P (2004). Pharmaceutical Microbiology. 7th ed. Blackwell Publishing Company, USA.
Fernandes Prabhavathi (2006). Antibacterial discovery – the failure of success? Nature Biotechnology, 24(12):1.
Finch R.G, Greenwood D, Norrby R and Whitley R (2002). Antibiotic and chemotherapy, 8th edition. Churchill Livingstone, London and Edinburg.
Hart C.A (1998). Antibiotic Resistance: an increasing problem? BMJ, 316:1255-1256.
Jayaramah R (2009). Antibiotic Resistance: an overview of mechanisms and a paradigm shift. Current Science, 96(11):1475-1484.
Mascaretti O.A (2003). Bacteria versus antibacterial agents: An integrated approach. Washington: ASM Press.
Mascaretti O.A (2003). Bacteria versus antimicrobial agents: An Integrated Approach. Washington: ASM Press.
Mazel D and Davies J (1998). Antibiotic Resistance: The Big Picture. In B. Rosen and S. Mobashery (Eds). Resolving the antibiotic paradox: progress in understanding drug resistance and developments of new antibiotics. New York: Plenum Press.
Russell A.D and Chopra I (1996). Understanding antibacterial action and resistance. 2nd edition. Ellis Horwood Publishers, New York, USA.
Sundsfjord A., Simonsen G.S., Haldorsen B.C., Haaheim H., Hjelmevoll S., Littauer P and Dahl K.H (2004). Genetic methods for detection of antimicrobial resistance. APMIS, 112:815-37.
Twyman R.M (1998). Advanced Molecular Biology: A Concise Reference. Bios Scientific Publishers. Oxford, UK.
Weaver R.F (2005). Molecular Biology. Third edition. McGraw-Hill Publishers, USA.
Willey J.M, Sherwood L.M and Woolverton C.J (2008). Harley and Klein’s Microbiology. 7th ed. McGraw-Hill Higher Education, USA.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.