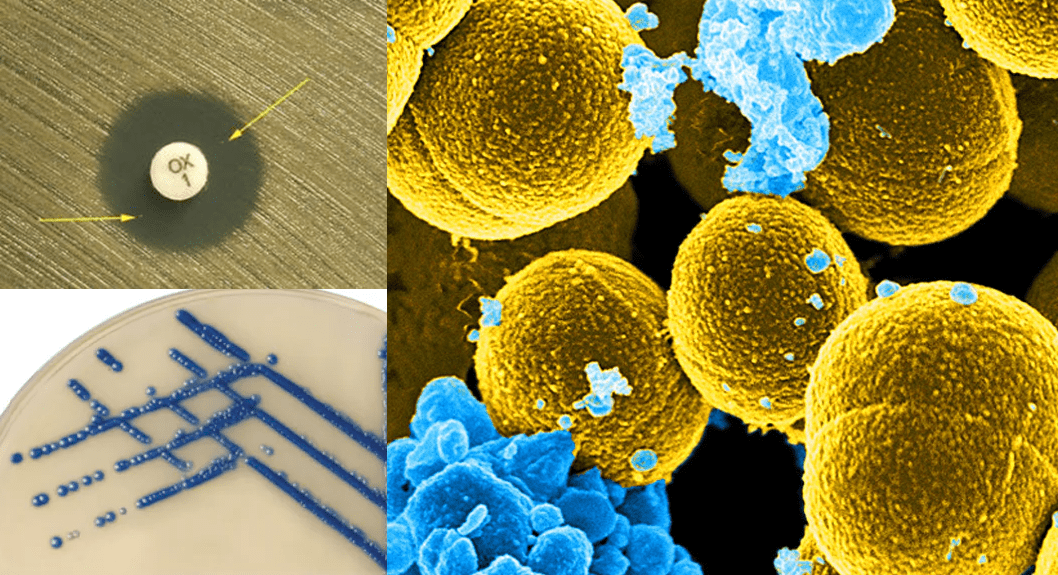
Antimicrobial agents, particularly antibiotics are the most significant class of pharmaceuticals and are one of the most influential medical inventions of the twentieth century. They have undeniably been a boon to human society in the fight against bacteria, saving millions of lives. Nonetheless, the number of infections caused by multidrug-resistant (MDR) bacteria is increasing across the world, and the threat of untreatable infections has been looming since the beginning of the 21st century. This is of serious public health importance because antibiotic resistance if allowed to continue may lunch the world into a post-antibiotic era – where no antibiotics will work or be efficacious against infectious diseases or their causative agents.
There has probably been a gene pool in nature for resistance to antibiotic as long as there has been for antibiotic production. This is because most microorganisms that are known to be antibiotic producers are resistant to even their own antibiotic. Antibiotic resistance in microorganisms is therefore a natural phenomenon, and it is spurred in microbes through selective pressure posed on pathogens by these agents (i.e. antimicrobials). Antibiotics no doubt have been able to stop the growth and kill many pathogenic microorganisms.
But microorganisms including bacteria, fungi, protozoa, and viruses have proven to be much more ingenious and adaptive to a pool of antimicrobial agents. They have developed resistance to some available antibiotics at an ever increasing pace than was previously imagined. Part of what have contributed to the dreaded increase in the emergence and spread of antibiotic resistant forms of microorganisms include bad practices on the part of some health practitioners when it comes to antibiotic prescription; and the mismanagement of some available antibiotics (especially the extended spectrum drugs) by both the health care practitioners and the general public have only help in exacerbating the status quo.
The history of antibiotic resistance is as old as the historical discovery and development of antimicrobial agents (antibiotic in particular) – since most cases of antibiotic resistance were discovered soon after the discovery and development of these antimicrobial agents. Antibiotic resistant bacteria were first discovered soon after the medicinal use of penicillin began. The first signs of antibiotic resistance were actually observed in 1940, five years before penicillin became commercially available to the public. In that year, the first observed bacterial enzyme (beta-lactamase) that destroyed penicillin was described.
This was the first observed evidence of bacterial resistance to an antibiotic action. Therefore, the history of antibiotic resistance coincided with the history of antibiotics themselves. The number of antibiotics belonging to various families, their varied mode of action and the number of bacteria in which antibiotic resistance has been documented suggests that, in principle, any microbe could develop resistance to any antibiotic. Antibiotic resistance is one of the biggest challenges that bedevil our health sector worldwide.
Resistance of microbes to antibiotics has been documented not only against antibiotics of natural and semi – synthetic origin, but also against purely synthetic compounds (such as the fluoroquinolones) or those which do not even enter the cells (such as vancomycin). And unfortunately, the discovery and development of newer antibiotics have not kept pace with the emergence and rate at which bacteria develops and mount resistance to antibiotics. Thus, the rate at which microbes are developing resistance to antibiotics is much faster than the rate at which the drugs are developed to curb the problem.
Pathogenic bacteria express a variety of mechanisms with which it evades the activity of potent antimicrobial agents including antibiotics, antifungal agents and antiviral agents amongst others. They use these mechanisms to remain active even in the face of antimicrobial onslaught; and this allows pathogenic organisms to acquire resistance genes which they propagate and transfer to susceptible microbes. Efflux pump mechanism, production of beta-lactamase enzymes and other antibiotic degrading-enzymes are some of the notable means with which pathogenic microorganisms uses to ward-off the action of antimicrobial agents directed towards them (Figure 1; Table 1).
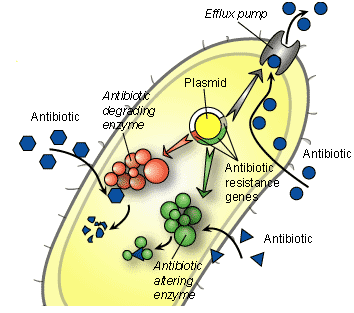
Table 1. Modes of action and resistance mechanisms of commonly used antibiotics
| Antibiotic class | Example(s) | Target | Mode(s) of resistance |
|---|---|---|---|
| β-Lactams | Penicillins (ampicillin), cephalosporins (cephamycin), penems (meropenem), monobactams (aztreonam) | Peptidoglycan biosynthesis | Hydrolysis, efflux, altered target |
| Aminoglycosides | Gentamicin, streptomycin, spectinomycin | Translation | Phosphorylation, acetylation, nucleotidylation, efflux, altered target |
| Glycopeptides | Vancomycin, teicoplanin | Peptidoglycan biosynthesis | Reprogramming peptidoglycan biosynthesis |
| Tetracyclines | Minocycline, tigecycline | Translation | Monooxygenation, efflux, altered target |
| Macrolides | Erythromycin, azithromicin | Translation | Hydrolysis, glycosylation, phosphorylation, efflux, altered target |
| Lincosamides | Clindamycin | Translation | Nucleotidylation, efflux, altered target |
| Streptogramins | Synercid | Translation | C-O lyase (type B streptogramins), acetylation (type A streptogramins), efflux, altered target |
| Oxazolidinones | Linezolid | Translation | Efflux, altered target |
| Phenicols | Chloramphenicol | Translation | Acetylation, efflux, altered target |
| Quinolones | Ciprofloxacin | DNA replication | Acetylation, efflux, altered target |
| Pyrimidines | Trimethoprim | C1 metabolism | Efflux, altered target |
| Sulfonamides | Sulfamethoxazole | C1 metabolism | Efflux, altered target |
| Rifamycins | Rifampin | Transcription | ADP-ribosylation, efflux, altered target |
| Lipopeptides | Daptomycin | Cell membrane | Altered target |
| Cationic peptides | Colistin | Cell membrane | Altered target, efflux |
References
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Bisht R., Katiyar A., Singh R and Mittal P (2009). Antibiotic Resistance – A Global Issue of Concern. Asian Journal of Pharmaceutical and Clinical Research, 2 (2):34-39.
Courvalin P, Leclercq R and Rice L.B (2010). Antibiogram. ESKA Publishing, ASM Press, Canada.
Denyer S.P., Hodges N.A and Gorman S.P (2004). Pharmaceutical Microbiology. 7th ed. Blackwell Publishing Company, USA.
Ejikeugwu Chika, Ikegbunam Moses, Ugwu Chigozie, Eze Peter, Iroha Ifeanyichukwu, and Esimone Charles (2013). Phenotypic Detection of Klebsiella pneumoniae Strains – Producing Extended Spectrum β-Lactamase (ESBL) Enzymes. Scholars Academic Journal of Biosciences. 1(1):20-23.
Ejikeugwu Chika, Umeokoli Blessing, Iroha Ifeanyichukwu, Ugwu Malachy, Esimone Charles (2015). Phytochemical and Antibacterial Screening of Crude Extracts from Leaves of Wonderful Kola. American Journal of Life Sciences. Special Issue: Microbiology Research, 3(2):5-8.
Ejikeugwu P.C., Ugwu C.M., Araka C.O., Gugu T.H., Iroha I.R., Adikwu M.U and Esimone C.O (2012). Imipenem and Meropenem resistance amongst ESBL producing Escherichia coli and Klebsiella pneumoniae clinical isolates. International Research Journal of Microbiology. 3(10):339-344.
Fernandes Prabhavathi (2006). Antibacterial discovery – the failure of success? Nature Biotechnology, 24(12):1.
Finch R.G, Greenwood D, Norrby R and Whitley R (2002). Antibiotic and chemotherapy, 8th edition. Churchill Livingstone, London and Edinburg.
Hart C.A (1998). Antibiotic Resistance: an increasing problem? BMJ, 316:1255-1256.
Jayaramah R (2009). Antibiotic Resistance: an overview of mechanisms and a paradigm shift. Current Science, 96(11):1475-1484.
Mascaretti O.A (2003). Bacteria versus antibacterial agents: An integrated approach. Washington: ASM Press.
Mascaretti O.A (2003). Bacteria versus antimicrobial agents: An Integrated Approach. Washington: ASM Press.
Mazel D and Davies J (1998). Antibiotic Resistance: The Big Picture. In B. Rosen and S. Mobashery (Eds). Resolving the antibiotic paradox: progress in understanding drug resistance and developments of new antibiotics. New York: Plenum Press.
Russell A.D and Chopra I (1996). Understanding antibacterial action and resistance. 2nd edition. Ellis Horwood Publishers, New York, USA.
Sundsfjord A., Simonsen G.S., Haldorsen B.C., Haaheim H., Hjelmevoll S., Littauer P and Dahl K.H (2004). Genetic methods for detection of antimicrobial resistance. APMIS, 112:815-37.
Twyman R.M (1998). Advanced Molecular Biology: A Concise Reference. Bios Scientific Publishers. Oxford, UK.
Weaver R.F (2005). Molecular Biology. Third edition. McGraw-Hill Publishers, USA.
Willey J.M, Sherwood L.M and Woolverton C.J (2008). Harley and Klein’s Microbiology. 7th ed. McGraw-Hill Higher Education, USA.
Discover more from Microbiology Class
Subscribe to get the latest posts sent to your email.