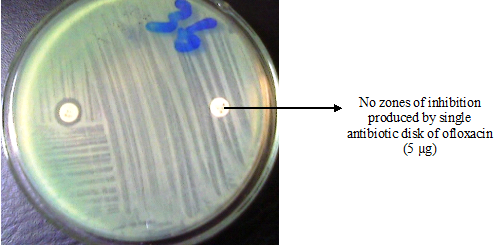
Antibiotic or antimicrobial resistance (AMR) is a phenomenon that occurs when bacteria are not killed or inhibited by usually achievable systemic concentration of an antibiotic (drug) with normal dosage schedule and/or fall in the minimum inhibitory concentration ranges of the drug in question. It is the ability of bacteria to resist the killing or inhibitory effects (onslaught) of an antibiotic.
Antibiotic resistant bacteria were first discovered soon after the first introduction of the first antibiotic and medicinal use of drugs particularly penicillin in clinical medicine for the treatment and management of infectious diseases. The first signs of antibiotic resistance were actually observed in the early 1940s, some few years before penicillin became commercially available to the general public for use in infectious diseases management and control. The first observed bacterial enzyme (beta-lactamase) that destroyed penicillin antibiotic was first described during this period.
Beta-lactamases are enzymes that hydrolyze and render inefficacious antibiotics that contain a 4-membered beta lactam ring such as penicillins and cephalosporins. These enzymes are usually secreted by microbes particularly bacteria in vivo or in vitro in response to an antimicrobial action, and as a way of surviving the onslaught of the antimicrobial agent or antibiotic targeted towards it. Today, there are plethora of beta-lactamase enzymes and even some mutated forms of these enzymes such as extended spectrum beta-lactamases (ESBLs) and metallo beta lactamases (MBLs) that confer on bacteria the ability to become multidrug resistant.
The discovery of bacterial enzymes (i.e. the beta lactamase enzymes) was indeed the first observed evidence of bacterial resistance to an antibiotic action. Therefore, it would not be far-fetched from the truth that the history of antibiotic resistance actually coincided with the history of antibiotic discovery. The number of antibiotics belonging to various families, their varied mode of action and the number of bacteria in which antibiotic resistance has been documented suggests that, in principle, any microbe could develop resistance to any antibiotic.
It is also not worthy that most microorganisms that are antibiotic producers are resistant to their own antibiotic; and this phenomenon actually gives impetus to the school of taught that microbial resistance is a natural process of microbial adaptation in their environment. Antibiotic resistance is one of the biggest challenges that bedevil our health sector worldwide, and this medical quagmire threatens our ability to effectively manage and treat some infectious diseases.
Microbial resistance to antibiotics and/or antimicrobial agents has been documented not only against antibiotics of natural and semi-synthetic origin such as the penicillins, but also against some purely synthetic compounds (such as the fluoroquinolones) or those which do not even enter the cells (such as vancomycin). And unfortunately, the slow pace in the discovery and development of novel antibiotics have not actually kept pace with the emergence and rate at which bacteria develops and mount resistance to some available antibiotics.
Penicillin (the first commercialized antibiotic) was serendipitously discovered by Alexander Fleming in 1928. It was not until 1945 that penicillin was distributed for use among the general public. This antibiotic was widely used in World War II to treat wounded soldiers of their surgical and wound infections; and penicillin was hailed as a “miracle drug or magic bullet due to its potent action in containing the excesses of bacterial infections as at the time. This set the stage for the discovery and development of several other antibiotics and/or antimicrobial agents including streptomycin, tetracycline and chloramphenicol capable of treating and bringing under control the excesses of some infectious diseases.
This is because the discovery and usage of antibiotics in infectious disease management has helped to reduce the rate of morbidity and mortality caused by infectious disease pathogens in human population. Nevertheless, some infectious diseases including but not limited to tuberculosis (TB), bacterial pneumonia, septicaemia, gonorrhea, wound infections and otitis media are now becoming recalcitrant to treat with some available antibiotics because the causative agents of these diseases are fast becoming resistant to some available antibiotic therapy.
These organisms have developed several novel ways and mechanisms that allow them to ward-off the antimicrobial onslaught of potent antimicrobial agents and/or antibiotics targeted towards them. Some available antimicrobial agents have been underused or misused in human medicine, agriculture and veterinary practice; and this development have allowed these microbes to develop resistance to these agents through selective pressure.
Antibiotic selective pressure is simply defined as the impact of antibiotic usage on a population of microorganisms – in which the microbes that are resistant to the antibiotic gain (or acquire) a survival advantage over those bacteria that are susceptible to the antimicrobial onslaught of the drug. The advantage gained by the resistant microbes is so unique because it allows them to take over the niche or habitat left by the antibiotic-susceptible bacteria (which were all killed by the antibiotic); and thus the resistant bacteria begins to spread in that particular environment even as it proliferates and ensures a continued reservoir of antibiotic-resistant bacteria.
This kind of development allows some chronic infections caused by drug-resistant microbes to persist and cause more harm to the individual. Microbial strains that harbour antibiotic resistance genes are usually more likely to clonally disseminate under some environmental conditions such as antibiotic selective pressure that encourages their dissemination within a given community.
Clonal dissemination of microbes refers to the spread of specific clones of an organism throughout a particular community. To prevent antibiotic selective pressure amongst bacterial populations and other microbes, it is critical to restrict and limit as much as possible antimicrobial usage especially for non-clinical purposes such as is obtainable in agricultural practices. Maintaining proper personal and environmental hygiene as well as good infection control practices in both the hospital and community settings are all important for the containment of antibiotic-resistant bacteria.
Antibiotic resistance in microbial pathogens should be timely and accurately detected as they emerge in order to avoid there spread within a given locality. And it is vital to also take necessary concerted efforts to contain there nefarious activities both within a hospital environment and in the community. However, the prevention of the problem through the rational use of available drugs is more vital to the detection of these organisms.
While the development of resistant strains of microorganisms is inevitable (since the process is part of microbial adaptation in the environment), the slack ways that antibiotics are administered and used in the hospitals and outside the hospital environment (especially in veterinary practice, livestock production and poultry production) has no doubt greatly exacerbated the rate at which drug resistant microbes emerge and spread.
Antibiotic resistance is simply defined as the ability of bacteria to resist the killing or inhibitory effects (onslaught) of an antibiotic. It is also the ability of microorganisms including bacteria, fungi, protozoa and viruses to resist (i.e. continue to live in the presence of an antimicrobial agent) the antimicrobial onslaught of a potent antimicrobial agent that is supposed to kill or inhibit its growth either in vivo or in vitro. Antibiotic resistance is a phenomenon that occurs when bacteria are not killed or inhibited by usually achievable systemic concentration of an antibiotic (drug) with normal dosage schedule and/or fall in the minimum inhibitory concentration (MIC) ranges of the drug in question.
Antibiotic resistance occurs when bacteria change in some way that reduces or eliminates the effectiveness of drugs or other agents designed to cure or prevent the infection. Thus the bacteria survive and continue to multiply causing more harm in the host taking the drug instead of abating the patient’s disease condition. Microorganisms harbor resistance genes and/or traits which encode various mechanisms that allow them to resist the killing or inhibitory effects of specific antibiotics directed towards them. These mechanisms also offer resistance to other antibiotics or antimicrobial agents of the same class and sometimes to several different antimicrobial classes. Resistant microorganisms survive exposure to a given antimicrobial agent and continue to multiply in the body, potentially causing more harm and spreading to other animals or people directly or indirectly.
Antimicrobial resistance is the ability of microbes, such as bacteria, viruses, parasites, or fungi, to grow in the presence of a chemical (drug) that would normally kill it or limit its growth. The terms antibiotic resistance and antimicrobial resistance can be synonymously used but with caution. The problem of antibiotic resistance is a vital topic and a global health problem that has received increasing attention over the last two decades. It is certainly not a new topic neither was it unpredictable. When antibiotic resistance occurs, it is the microbe (bacterium) that is resistant, not the antibiotic nor the patient or host taking it. Species of bacteria that are normally resistant to penicillin for example, can develop resistance to these drugs either through mutation (vertical transmission) or through the acquisition of resistance genes from other bacteria (horizontal transmission).
This dual means of acquiring resistance (i.e. the vertical and horizontal transmission) explains why the resistance trait can spread rapidly and replace a previously drug-susceptible population of bacteria. Antibiotic resistance is on the rise threatening our ability to treat infectious disease. Antibiotic resistance development is a natural process of microbial adaption in their natural environment; and this phenomenon often leads to a limited lifespan of antibiotics used for the treatment of human illnesses. It is noteworthy that all types of microbes (including bacteria, fungi, protozoa and viruses) have the ability to naturally develop resistance to antimicrobial agents created to kill or inhibit their unwanted growth since this process (i.e. microbial resistance) is a natural phenomenon that allows the organism to survive adverse conditions (including exposure to antibiotics) in its environment.
In the end, the exposed microbes become drug-resistant organisms – capable of warding-off the action of antimicrobial agents or antibiotics. However, the unnecessary and inappropriate use of antibiotics and other antimicrobial agents favours the emergence and spread of resistant microbes in both the community and hospital settings. The use of antibiotics (either rationally or irrationally) is the single most important factor leading to antibiotic resistance around the world.
Since simply using antibiotics creates resistance in the microbial world, it is therefore vital that we only use them wisely especially in the area of infectious disease management.
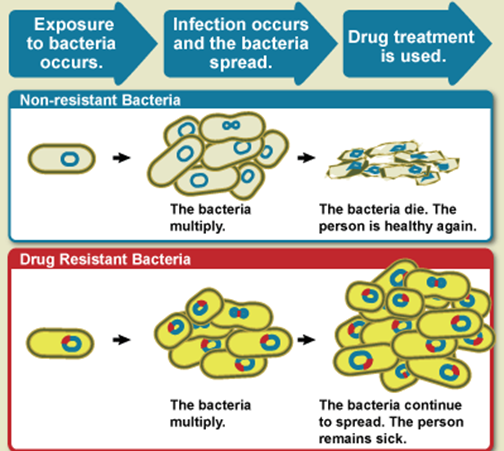
Non-resistant bacteria multiply, and upon proper drug treatment, the non-resistant bacteria die. Nevertheless, drug resistant bacteria multiply as well during this process of treatment, but upon drug treatment, the drug resistant bacteria continue to multiply and spread within the host (Figure 1). Some common and life-threatening infectious diseases such as TB are becoming difficult or even impossible to treat because of drug-resistant bacteria. Microbial infections caused by resistant microorganisms are often difficult to treat, and such infections usually require high cost and sometimes toxic alternatives to get rid of them from the body. Moreover, patients infected with drug-resistant bacteria often spend long time in the hospital; and long hospitalization makes such individuals prone to acquiring nosocomial (hospital-acquired) infections.
Antimicrobial resistance makes it difficult and more expensive to treat many common microbial infections as aforementioned; and this causes delays in effective treatment or, in worst cases, inability to provide treatment at all – since the resistant organisms are often not susceptible to some commonly available antimicrobial agents or antibiotics. Antimicrobial resistance is a serious public health problem that strikes at the core of infectious disease control and management; and if not handled now and carefully, it has the potential to halt, and possibly reverse the progresses gained since the development and use of antibiotics in clinical medicine for treatment of human diseases.
Though antimicrobial or antibiotic resistance is a natural response of microbes to surviving harsh environmental conditions as aforesaid, the problem of antibiotic resistance can still be contained through the careful and appropriate use of available antibiotics or antimicrobial agents. Drug resistant microbes can end up in our food from animal sources in several ways. All animals carry bacteria in their gastrointestinal tract (GIT); and these microbes in the gut of these animals play several beneficial roles in the digestion of the food that they eat. Animals including livestock and poultry birds are often given antibiotics as growth promoting agents. In some cases, the feed of these animals are usually mixed or compounded with antibiotics – which act as growth promoting agents.
The administered antibiotics will only kill susceptible bacteria while the resistant bacteria will survive and continue to multiply. When food animals are slaughtered and processed, these resistant bacteria can contaminate the meat or other animal products. And the resistant bacteria can get into the environment such as water supplies through animal effluents. The resistant bacteria in the feacal matter of the animal may spread to farm products or crops that are irrigated with feacally-contaminated water. Drug resistant microbes can spread from person to person, or from the non-human sources in the environment such as animals to humans, and this process allows these microbes to spread in both the hospital and community settings.
There are several direct routes by which people can get antibiotic-resistant bacteria that develop in industrial food animal production. When meat or animal products are not handled or cooked properly, drug-resistant bacteria in the meat can spread to humans through consumption (Figure 2). Humans can get drug-resistant bacteria from animals through improper handling or consumption of inadequately cooked contaminated meat or other animal products such as milk. Direct body contact with infected farm workers and/or meat processors, and veterinary doctors is another important route via which resistant bacteria can spread from animals to humans. Resistant bacteria can also spread through drinking contaminated surface or ground water and eating contaminated crops especially those irrigated with feacally-contaminated water as aforesaid.
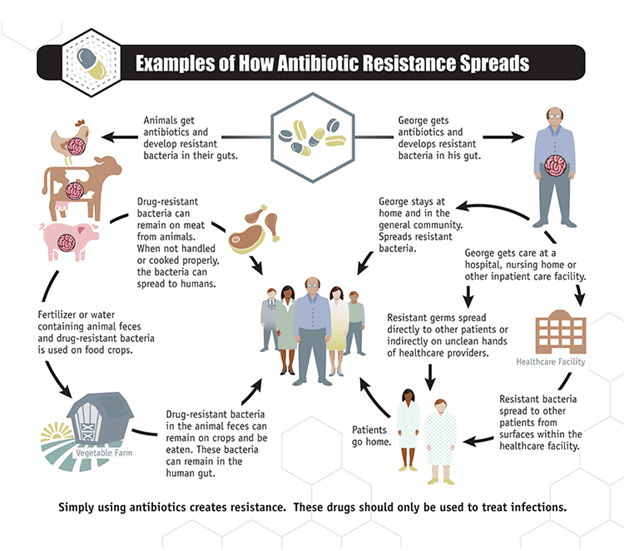
Antibiotic resistant bacteria can also be spread or contacted by contaminated air that is vented from concentrated animal housing or is released during animal transport; and people who live close to animal farms are at high risk of contracting such microbes. It is now becoming clearer that the misuse of antibiotics especially as growth promoting agents in livestock and poultry production is an important factor of the antibiotic resistant problems we are facing today. Thus it is critical to only use antibiotics and/or antimicrobial agents with caution especially when there usage in animals is anticipated.
References
Arora D.R (2004). Quality assurance in microbiology. Indian J Med Microbiol, 22:81-86.
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Axelsen P.H (2002). Essentials of antimicrobial pharmacology. Humana Press, Totowa, New Jersey, USA. Al-Jasser A.M (2006). Extended – Spectrum Beta – Lactamases (ESBLs): A Global Problem. Kuwait Medical Journal, 38(3):171-185.
Bisht R., Katiyar A., Singh R and Mittal P (2009). Antibiotic Resistance – A Global Issue of Concern. Asian Journal of Pharmaceutical and Clinical Research, 2 (2):34-39.
Block S.S (2001). Disinfection, sterilization and preservation. 5th edition. Lippincott Williams & Wilkins, Philadelphia and London.
Cars O and Nordberg P (2005). Antibiotic resistance: The faceless threat. International Journal of Risk & Safety in Medicine, 17 (3/4): 103-110.
Cowan M.M (1999). Plant products as antimicrobial agents. Clinical Microbiology Reviews., 564-582.
Denyer S.P., Hodges N.A and Gorman S.P (2004). Pharmaceutical Microbiology. 7th ed. Blackwell Publishing Company, USA.
Ejikeugwu Chika, Ikegbunam Moses, Ugwu Chigozie, Eze Peter, Iroha Ifeanyichukwu, and Esimone Charles (2013). Phenotypic Detection of Klebsiella pneumoniae Strains – Producing Extended Spectrum β-Lactamase (ESBL) Enzymes. Scholars Academic Journal of Biosciences. 1(1):20-23.
Ejikeugwu Chika, Iroha Ifeanyichukwu, Adikwu Michael and Esimone Charles (2013). Susceptibility and Detection of Extended Spectrum β-Lactamase Enzymes from Otitis Media Pathogens. American Journal of Infectious Diseases. 9(1):24-29.
Ejikeugwu Chika, Iroha Ifeanyichukwu, Adikwu Michael and Esimone Charles (2013). Susceptibility and Detection of Extended Spectrum β-Lactamase Enzymes from Otitis Media Pathogens. American Journal of Infectious Diseases. 9(1):24-29.
Finch R.G, Greenwood D, Norrby R and Whitley R (2002). Antibiotic and chemotherapy, 8th edition. Churchill Livingstone, London and Edinburg.
Joslyn, L. J. (2000). Sterilization by Heat. In S. S. Block (Ed.), Disinfection, Sterilization, and Preservation (5th ed., pp. 695-728). Philadelphia, USA: Lippincott Williams and Wilkins.
Lai P.K and Roy J (2004). Antimicrobial and chemopreventive properties of herbs and spices. Curr. Med. Chem, 11 (11): 1451–1460.
Livermore D.M (2004). The need for new antibiotics. Clinical Microbiology & Infection, 4(10): 1-9.
Mascaretti O.A (2003). Bacteria versus antibacterial agents: An integrated approach. Washington: ASM Press.
Nally J.D (Ed.) (2007). Good manufacturing practices for pharmaceuticals. Sixth edition. Informa Healthcare USA, Inc, New York.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.