An antibiotic has to go through a number of steps in order to exert its antibacterial action in vivo. They have to come into contact with the host taking them before ever their antibacterial and/or antimicrobial properties can be dissipated especially in the aspect of treating and abating a given disease condition or process. To start with, the antibiotic has to enter the host cell, and once inside the cell, the antibiotic has to remain stable and accumulate to killing or inhibitory concentrations enough to deactivate invading microorganisms (or bacteria).
In some cases, the antibiotic need to be activated to an active form and finally it has to locate and interact with its target(s) in order to exert its action. Usually, microorganisms including bacteria, fungi, protozoa and viruses have specific target sites on their cell walls or cell membranes upon which antibiotics or antimicrobial agents must first bind to in order to initiate their antimicrobial onslaught. Any alteration in any one or more of these processes can render the cells of the bacteria resistant to the antibiotic directed against it.
This is what happens whenever a bacterium mounts resistance to antibiotics which was supposed to inactivate their degrading effects to the host, hence the need to acquaint ourselves with the mechanisms of acquiring resistance by bacteria. In addition to this, the increased dissemination and prevalence of resistance is an outcome of natural selection and should be viewed as an expected phenomenon of the “Darwinian” biological principle of “survival of the fittest”. In any large population of bacteria, a few bacterial cells will usually be present which possesses traits that enable them to survive in the presence of harmful substances such as antibiotics.
This exemplifies the ability of the bacteria to fade-off or evade the action of the antibiotic directed against them. Susceptible organisms (i.e. those lacking the advantageous traits like antibiotic resistance genes) will be eliminated in the face of the antimicrobial onslaught, leaving behind the remaining resistant population of bacteria. With long time exposure to a given array of antibiotics in a given environment, the bacteria communities will change dramatically, with more resistant organisms increasing in proportion.
The bacteria is said to have acquired resistant traits or become resistant through selective pressure due to undue exposure to the drugs. In such scenarios of antibiotic resistance, the antibiotics to which the bacteria are resistant to may not be effective enough to treat what was once and easily treatable infection.
The bacteria are now unaffected by the antibiotic because of selective pressure posed on them by initial, “unnecessary”, and longtime antibiotic usage or exposure in a particular population or environment. Some of the notable mechanisms employed by bacteria to mount resistance against antimicrobial agents (antibiotics) are as follows:
ANTIBIOTICS RESISTANCE BY INFLUX-EFFLUX SYSTEMS
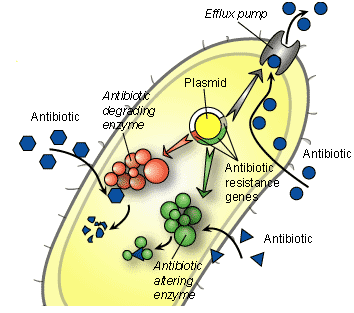
Certain bacteria can often become resistant to antimicrobial agents or antibiotics through a mechanism known as “efflux”. Efflux pumps are pumps found in bacteria cells. These pumps help them to export antimicrobial agents and/or antibiotics and other chemical compounds out of the bacterial cell. The antibiotics enter the bacteria through chemical channels called “porins” found on the bacterial cell membranes. However, some resistant bacteria with the influx-efflux mechanisms uses this system to pump out antimicrobial agents from the cell – in order to prevent the intracellular accumulation of the drug required to kill or inhibit an important metabolic process in the target bacterium.
By actively pumping out the antibiotic and other harmful substances out of the cell, the efflux pumps prevents the intracellular accumulation of the antimicrobial agent that is necessary to exert optimal antibacterial activity inside the bacterial cell. Increased expression of the efflux pump mechanisms in bacteria can result in antibiotic resistance in bacterial population. Bacterial cells have an inherent (natural) capacity to restrict the entry of small molecules (e.g. antibiotics) that destabilizes its internal metabolism and interferes with the normal growth and developmental process of the organism.
This is what the cell wall and outer cell membranes in both Gram positive and Gram negative bacteria respectively do. The ability of bacteria to restrict the entry of harmful materials into their internal environment is more pronounced in Gram negative bacteria unlike in Gram positive bacteria which are devoid of “outer membrane” that the former possesses. The “outer membrane” is a first-line defense (protection) mechanism in Gram negative bacteria, and its absence in Gram positive bacteria is the reason why Gram positive bacteria is highly sensitive to antibiotics than their Gram negative counterparts.
This is because there is no form of security or outer covering to protect the peptidoglycan of Gram positive bacteria as is applicable in Gram negative bacteria with outer membrane. The “influx – efflux” system in bacteria has to do with the entry and partial accumulation of harmful substances like antibiotics within the cytoplasm of a bacterium and the subsequent exit or removal of these harmful substances from the bacterial cell through efflux pumps.
The “efflux system” in bacterial cell pumps out the antibiotics that finally made their way into the cytoplasm of the bacterium, thereby preventing their intracellular accumulation. The most well studied efflux system is in Escherichia coli and with this mechanism in place; bacteria can easily mount resistance to antibiotics directed against them.
ANTIBIOTIC RESISTANCE BY CHEMICAL ALTERATION OF ANTIBIOTICS IN VIVO
Antibiotics are expected to exert killing or inhibitory effects on their target pathogenic microorganisms when in vivo. However, some resistant microbes have developed mechanisms through which they alter the chemical composition or structural/functional groups of antibiotics – so that the antimicrobial agent will be rendered inefficacious for therapy in vivo. Chemical alteration of antibiotics is thus one major mechanism via which bacteria can become resistant to an antibiotic.
Some antibiotics need to be activated in vivo before ever they can educe (bring out) their antibacterial activity against a target pathogen to which they were meant to attack and kill or inhibit its growth. Antibiotics whose functional group are deactivated or altered in vivo by bacteria are usually activated in vivo by being reduced by a specific enzyme or gene; and only then can such drug be able to elicit their biological properties in vivo. For example, antibiotics in the Nitrofuran family such as nitrofurantoin (notable for their usage in treating urinary tract infections, UTIs) are reduced by cellular reductase enzymes encoded by specific genes in the target organism; and any mutation in these genes can eventually lead to a nitrofuran resistance.
More so, some pathogens produce enzymes such as beta-lactamase enzymes that alter the chemical structure and/or biological function of some antibiotics. Thus, the chemical alterations of some antibiotics such as antibiotics in the beta-lactam group like the penicillins and cephalosporins in vivo by antibiotic-degrading enzymes (e.g. beta-lactamases) can inactivate the biological activity of these antibiotics, thereby leading to resistance.
ANTIBIOTIC RESISTANCE DUE TO TARGET ALTERATIONS
Microorganisms including bacteria and fungi have specific target site(s) for drug-binding on their cell wall or cell membranes. Antibiotic resistance in bacteria could develop as a result of alteration in any of these target site(s); and this prevents the drug from binding and carrying out its notable antimicrobial activity either in vivo or in vitro. Most pathogenshave theability to alter target sites(s) of antibiotics in their cell.
These alterations as aforementioned in the target of the drugs on the organism occur in such a way that the action of the antibiotic on the target pathogen is countered or prevented. Alteration of the penicillin-binding-proteins (PBPs) in bacteria result in antibiotic resistance to some classes of antibiotic especially the beta-lactam drugs such as the penicillins that have high affinity for the PBPs. The penicillin-binding-proteins (PBPs) are transpeptidases which catalyze the cross-linking reaction between two stem peptides – N-acetyl muramic acid (NAM) and N-acetyl glucosamine (NAG).
And each of these molecules is linked to the peptidoglycan backbone – which is the major component of bacterial cell wall. The reaction that catalyzes the cross-linking of NAM and NAG is known as transpeptidation reaction; and this reaction leads to the formation of peptidoglycan layer – that gives rigidity to the bacterial cell wall. Penicillin and other beta-lactam drugs exerts their antibacterial activity by binding to the PBPs, thereby preventing the cross-linking of N-acetyl-muramic acid and N-acetyl-glucosamine that will eventually lead to the formation of a very rigid bacterial cell wall.
But alterations in this important drug target sites on the pathogen (i.e. the PBPs) can make the organism to resist antimicrobial action from beta-lactam drugs. The alterations in drug target sites could also be mutational – in which case a mutation in the target site could prevent the binding of the drug.
ANTIBIOTIC RESISTANCE DUE TO LOSS OF DRUG ENTRY POINTS (PORIN CHANNELS)
Porin channels or porins are proteins that assemble to form a complex that acts as channel where molecules including substrates and antibiotics can pass through the cell of an organism. Porins are found in the outer membranes of Gram-negative bacteria. Porin channels are also found in the mitochondria of eukaryotic organisms and the chloroplasts of plant cells, where they form ion-selective channels for the entry of small hydrophilic molecules including antibiotics and/or antimicrobial agents.
Porins are involved in the exchange of nutrients over the outer membrane (OM) of Gram-negative bacteria but they are also involved in the pathogenesis of the organism since porin channels could influence the development of antibiotic resistance through the modifications of the properties of the OM lipid barrier of a bacterium. Most importantly, the porin channels prevent the entry of noxious molecules such as antibiotics into the bacterial cell.
The outer membrane of Gram-negative bacteria provides a formidable barrier that must be overcome before drug penetration into the cell. This formidable barrier of the Gram negative cell wall (unlike the less-formidable barrier of the Gram positive cell wall) also accounts for the high levels of antibiotic resistance experienced in Gram negative bacteria including Klebsiella species, Escherichia coli, and Pseudomonas aeruginosa. Alterations or loss of drug entry portals such as the porin channels on bacteria could lead to antibiotic resistance. This could occur when antibiotics find it difficult to locate their portal of entry into the internal environment of their target organism.
Porins influence on the emergence of antibiotic resistant strains of pathogenic bacteria especially when drugs fail to penetrate their target organisms. Small hydrophilic drugs, such as the beta-lactams and non-beta-lactams including tetracycline, chloramphenicol and fluoroquinolones, use the porin channels to gain access to the internal environment of their target pathogen. However, some hydrophobic drugs such as the macrolides (e.g. erythromycin and azithromycin) diffuse across the lipid bilayer of the organism in order to enter the cell. Any decrease in the ability or rate of entry of antibiotics into the cell of an organism can lead to resistance.
Microbial resistance could emerge when a decrease in the rate of drug entry is generated in bacteria; and this usually happens when the porin channels for drug entry are either modified or altered via mutation. The emergence of drug-resistant strains of microbes in a large number of bacterial species due to the modifications or alterations of their lipid or protein composition of the organism’s OM highlights the importance of the OM barrier in antibiotic sensitivity. Mechanisms affecting the barrier properties of the OM through the modification of expression and/or function of the porin channels of bacteria have a huge impact on the sensitivity of Gram-negative bacteria to antibiotics.
ANTIBIOTIC RESISTANCE DUE TO NON-HERITABLE STATES OF BACTERIA
Microorganismsincluding bacteria and fungi have some non-heritable physiological states which are inherent with the organisms; and which aid or assist them to evade antimicrobial onslaughts of antimicrobial agents and/or antibiotics. These physiological states of microbes are not transferable like resistant plasmids and transposons that carry resistant traits and thus could be transmitted from one organism to another. The non-heritable form of antibiotic resistance posed by bacteria to antibiotics has to do with some physiological states in which bacteria exist in, and which are not heritable or transferred by other organisms.
These non-heritable physiological states of bacteria include the biofilm states of bacteria, swarming states of bacteria, and the persistence states of microbes. Microbes assume these non-heritable states in order to render bacteria or a community of organisms insensitive to the antimicrobial onslaughts of antibiotics. The physiological states of microbes are expressed in a transient state, and they are reversible and non-heritable in nature. In such states, bacteria are said to be antibiotic tolerant (i.e. they continue to thrive in the presence of potent antimicrobial activity).
The persistence state in bacteria is characterized by a state in which bacteria exist in a small fraction of slow or non-growing, antibiotic tolerant cells called persisters. These persisters (which are antibiotic-tolerant states of bacteria) exist in this form and remain insensitive to harmful substances such as antibiotics. Biofilms are organized microbial structures that consist of layers of microbial cells associated with either a biotic or abiotic surface. In biofilm states, bacterial cells of several species are embedded in a self-produced exo-polysaccharide matrix that is usually made up of lipopolysaccharides.
Biofilms form when bacteria adhere to surfaces in aqueous environments and begin to excrete a slimy, glue-like substance that can anchor them to all kinds of materials or surfaces including medical implant devices such as respirators and urine catheters. When bacteria come together, they form a biofilm-state – which allows the organisms to communicate via a process known as quorum sensing. Bacteria that engage in quorum sensing communicate their presence by emitting chemical messages or molecules that their fellow infectious agents in the environment are able to recognize.
This mechanism can allow the organisms in the quorum or biofilm-state to be resistant to untoward effects in their environment including antimicrobial action. Biofilm structures are highly organized and they permit the transport of nutrients and metabolic wastes in and out of the matrix structure. Biofilms have a very high tolerance to high concentrations of antibiotics.
Swarming is a form of multicellularity in many bacterial species such as Proteus species, and it is characterized by the migration of highly differentiated bacterial cells (swarm cells) on semi-solid surfaces. This gives them some form of protection from antibiotics as, they are known to remain in close contact with one another and they also migrate as a group in this form – thus dodging antimicrobial activity.
References
Arora D.R (2004). Quality assurance in microbiology. Indian J Med Microbiol, 22:81-86.
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Axelsen P.H (2002). Essentials of antimicrobial pharmacology. Humana Press, Totowa, New Jersey, USA. Al-Jasser A.M (2006). Extended – Spectrum Beta – Lactamases (ESBLs): A Global Problem. Kuwait Medical Journal, 38(3):171-185.
Bisht R., Katiyar A., Singh R and Mittal P (2009). Antibiotic Resistance – A Global Issue of Concern. Asian Journal of Pharmaceutical and Clinical Research, 2 (2):34-39.
Block S.S (2001). Disinfection, sterilization and preservation. 5th edition. Lippincott Williams & Wilkins, Philadelphia and London.
Cars O and Nordberg P (2005). Antibiotic resistance: The faceless threat. International Journal of Risk & Safety in Medicine, 17 (3/4): 103-110.
Ejikeugwu Chika, Iroha Ifeanyichukwu, Adikwu Michael and Esimone Charles (2013). Susceptibility and Detection of Extended Spectrum β-Lactamase Enzymes from Otitis Media Pathogens. American Journal of Infectious Diseases. 9(1):24-29.
Finch R.G, Greenwood D, Norrby R and Whitley R (2002). Antibiotic and chemotherapy, 8th edition. Churchill Livingstone, London and Edinburg.
Joslyn, L. J. (2000). Sterilization by Heat. In S. S. Block (Ed.), Disinfection, Sterilization, and Preservation (5th ed., pp. 695-728). Philadelphia, USA: Lippincott Williams and Wilkins.
Lai P.K and Roy J (2004). Antimicrobial and chemopreventive properties of herbs and spices. Curr. Med. Chem, 11 (11): 1451–1460.
Livermore D.M (2004). The need for new antibiotics. Clinical Microbiology & Infection, 4(10): 1-9.
Mascaretti O.A (2003). Bacteria versus antibacterial agents: An integrated approach. Washington: ASM Press.
Nally J.D (Ed.) (2007). Good manufacturing practices for pharmaceuticals. Sixth edition. Informa Healthcare USA, Inc, New York.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.