Replication is defined as the process in which a cell divides to make copies of its genome or itself. Cell division or replication in viruses is different from what is obtainable in other microbes such as bacteria that mainly replicates by binary fission. Replication in viruses only occurs inside a suitable host, and in such cases the virus redirects the metabolic and cellular machinery of its host cell to support the replication of the virus in vivo.
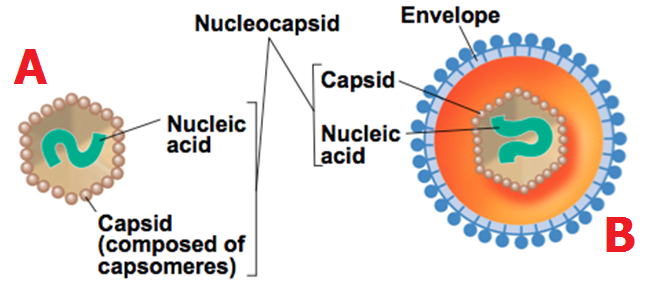
Viral genomes (DNA or RNA) are usually introduced into the host cell through infection, and once they have gained entrance the virus ensures that the host cell is overwhelmed to produce the different viral components (e.g. capsid and nucleic acid) required to generating new virions that will be released to continue the infection process.
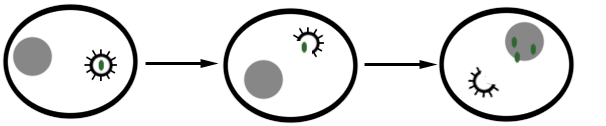
Viral replication usually involves several phases from the point of attachment of the virus to its host cell and the release of new virions from the host cell. Attachment, penetration, uncoating, expression of viral nucleic acid, biosynthesis of viral components, assembly and the release of mature and complete virions are the main stages that characterize the replication of viruses in a host cell (Figure 1).
ATTACHMENT STAGE OF VIRAL REPLICATION
As aforementioned, viruses usually gain entry into their host cell through infection. Attachment or adsorption is the first step in viral replication; and this stage is critical because without it the infecting virus cannot gain entry into its target host cell. In adsorption (attachment), the infecting virus attaches to specific receptors on the cell membrane of its target host cell through its capsid or surface proteins and this interaction between the infecting virus and the target host cell is vital for viral entry.
Absence of a particular receptor site on the host cell membrane that the infecting virus can recognize means that the infecting virus will not attach and this ultimately prevent infection because the infecting virus cannot attach. One of the major aims of an infecting virus is to replicate its genome especially in a host cell, and in order to achieve this the virus must find a way to first of all enter the target host cell and then takeover its metabolic and cellular machinery to manufacture its own viral components so that new virions can be generated and released.
But this cannot be possible if the infecting virus fails to attach itself first to specific protein molecules (inclusive of other lipoproteins, carbohydrates and glycoproteins) found on the cell or plasma membrane of the target host cell.
The first step involved in the replication of a virus is the attachment or adsorption of the infecting virus to the surface proteins found on the surface of its host cell. The protein molecules found on the capsid of the virus interact specifically with the surface receptors on the host cell; and this facilitates the entry or penetration of the infecting virus into the cell. After entry, the infecting virus uncoats and releases its nucleic acid genome (DNA or RNA) from its nucleocapsid or capsid. The released nucleic acid genome takes over the cellular machinery of the host cell and starts expressing its own genetic makeup. The expression of viral nucleic acid genome within the host cell leads to the biosynthesis of specific viral proteins and viral nucleic acids required for the assembling of new virions. Newly synthesized virions must be packaged into a complete virion or viral particle before it can be released from the host cell to infect new cells; and this is prerequisite for the infection of new cells within the organism. After proper assembling or packaging, the newly synthesized virions are released from the host cell through cell lysis (for naked viruses) or through budding (for enveloped viruses).
These receptors are unique and specific in nature, and viruses usually have several multiple protein sites that can bind to host cell receptors in a specific fashion in order to facilitate their entry into the host cell. Viral pathogenesis (i.e. the ability of a virus to cause infection in its host organism or cell) is largely dependent on the ability of the infecting virus to attach to receptors on the target host cell membrane; otherwise there will be no initiation of infection because viral entry will not be facilitated if the infecting virus fails to adsorb or attach to a specific receptor expressed the surface of a susceptible host cell.
It is noteworthy that susceptible host cells that fail to express specific receptors for viral attachment cannot be infected by the infecting virus; and the host cell can also become resistant to the infecting virus in cases where the target receptor on the surface of the host cell becomes mutated and altered in such a way that it cannot be recognized. The host cell is said to be resistant in such scenarios. However, some mutated viruses can still come up and attach to the mutated cell surface receptors. The cell surface receptors on the host cell generally determine whether the host cell will be infected or not by a particular infecting virus.
PENETRATION (ENTRY) STAGE OF VIRAL REPLICATION
Penetration is the next stage that follows a successful viral attachment in the viral replication cycle. Viruses usually penetrate or enter their target host cell after attachment through a biological process known as endocytosis. Endocytosis is simply defined as the process in which a virus is taken intact into a host cell. Viral entry into their host cell is critical because viral replication can only occur within a particular host cell and not outside it.
The mode of penetration of the infecting virus is usually different depending on whether the virus is a naked virus or an enveloped virus. However, the main aim of this stage is to ensure that the infecting viral genome penetrates the host cell so that it can be replicated. In naked or non-enveloped viruses, the plasma membrane of the susceptible host forms an endosome or endosomal cavities that enclose the infecting virus and this facilitate its entry through endocytosis into the host cell.
Enveloped viruses usually uncoat at the plasma membrane and this facilitates the entry of the virion content into the host cell. The fusion of the lipid envelop of enveloped viruses with the plasma membrane of the susceptible host cell facilitates the entry of the enveloped viruses into the host cell. Nonetheless, endocytosis is the main process via which the naked viruses and enveloped viruses enter their host cells.
UNCOATING STAGE OF VIRAL REPLICATION
Uncoating is another key step in the viral replication cycle. This is because it is at this stage that the viral nucleic acid genome (DNA or RNA) is released from the capsid or nucleocapsid of the infecting virion. This important stage of viral replication cycle can be inhibited by the host’s defense mechanisms such as the release of enzymes (e.g. lysozymes) that inactivate the pathogenic potential of the infecting virus prior to uncoating.
During uncoating, the viral nucleocapsid or capsid and other associated protein molecules is physically separated from the genome or removed in order to expose the viral genome which takes over the cellular and metabolic machinery of the host cell for the commencement of viral replication and the production of new viral components. At this stage of uncoating, the infecting parent virus losses its infectivity and thus it can longer be infectious since its protein coat or capsid has been physically separated from its genome.
EXPRESSION OF VIRAL NUCLEIC ACID AND BIOSYNTHESIS OF VIRAL COMPONENTS
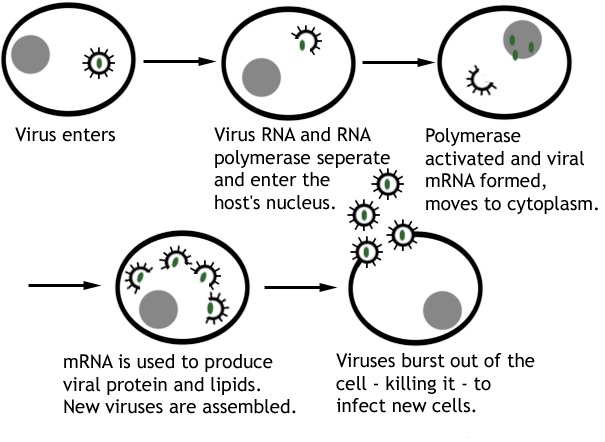
After uncoating as aforementioned, the viral nucleic acid genome (DNA or RNA) becomes expressed within the host cell and this stage usually follows the central dogma of molecular biology in which DNA is transcribed to mRNA that is later translated to specific proteins. Specific mRNA must be transcribed from the viral genome (e.g. DNA) for the successful expression and duplication of the genetic information of the infecting virus.
DNA replication mainly occurs in the nucleus of the infected host cell for DNA viruses; while for RNA viruses (whose genome is mainly mRNA in form), the viral RNA is mainly replicated in the cytoplasm of the cell. However, some DNA viruses (e.g. poxviruses) can still replicate their genome in the cytoplasm; and this variation is observable in some RNA viruses (e.g. retroviruses) that replicate their genome in the nucleus of the infected host cell.
Following infection, it is critical to produce new copies of the viral genome inside the host cell and this is usually accompanied by the synthesis of viral proteins as shall be highlighted later. An understanding of the replication mechanisms of infecting viruses (DNA and RNA viruses inclusive) is vital to the development of novel drugs that can interfere with and inhibit the pathogenic or virulent capabilities of pathogenic viruses especially in viral disease conditions.
The synthesis of virus nucleic acid and proteins is vital to viral replication, and this is carried out by the metabolism of the host cell as directed by the infecting virus. New copies of viral genome and specific viral proteins must be synthesized in the infected host cell prior to the formal replication of the said virus. The replication of a particular virus is usually directed by the orientation or structure of its genome. Generally, the messenger ribonucleic acid (mRNA) of a virus is defined as “positive sense” (designated with the symbol ‘+’) while its complementary strand is defined as “negative sense” (designated with the symbol ‘—’).
This implies that a nucleic acid strand that has the same sequence as mRNA is designated (+) strand while the one with a complementary strand is designated (—) strand. Andsince all viruses (both DNA and RNA viruses) must express their genes or genetic makeup as functional mRNA molecules early enough in the course of their infection or disease development in vivo (i.e. within a living host cell), the mRNA (which is unique in carrying and decoding the genetic information for protein synthesis from the gene) of the infecting virus must always be in preference to the host’s mRNAs potential viral proteins must be synthesized for the coupling of new virions.
Viruses as aforementioned are microbes that are composed mainly of nucleic acids and protein molecules and envelopes in the case of enveloped virus. The mRNA is responsible for the translation of genetic information it encodes (especially for the synthesis of specific viral proteins). It converts the genetic information it is carrying into a sequence of polypeptide in order to form a particular protein molecule; and this phenomenon is of utmost importance to virus because if the viral capsid (which is proteinous in nature) is not well synthesized, the viral genome will not be well packaged or assembled; and this generally exposes the virus to destruction by the host immune mechanisms and by other antiviral agents.
Thus, in order to direct the host cell’s translational machinery to make viral proteins; it is critical that the correct viral mRNAs which are designated as positive sense or negative sense depending on the genomic orientation of the virion are always available and well aligned to carry out this important genetic transformation. The phrases ‘sense’ and ‘strand’ are used synonymously in describing the genomic orientation of viruses. It is noteworthy that some viruses can have both the negative and positive strand, and for such viruses; their genomic orientation is said to be ambisense in nature.
Different viral families inclusive of DNA and RNA viruses have different genomic orientation that can either be single-stranded(ss) or double stranded (ds); and the polarity of their mRNA can either be positive sense, negative sense or ambisense as the case may be. These variations or features are critical in understanding the replication pattern of each of these viruses that make up the different viral families; and because of the differences in the transcription of the genomes of DNA viruses and RNA viruses during viral replication, it is important to use these designations as aforementioned in the discussions of viral replication for clarity.
The replication pattern of a particular virus is usually determined by the structural orientation and makeup of its genome. For example, viruses with single-stranded positive sense RNA (ss+RNA) genome make use of their positive RNA (+RNA) as mRNA during transcription. These viruses uses the ribosomes and enzymes of the host cell to decode the information encoded in the +RNA genome of the virus during translation to produce protein molecules (e.g. RNA-dependent RNA polymerase).
RNA-dependent RNA polymerase is responsible for transcribing the RNA genome of RNA viruses into RNA genomes so that such viruses can be further replicated. Eukaryotic cells do not possess enzymes for the replication of RNA genomes; and thus the infecting virus must provide replicase enzymes such as the RNA-dependent RNA polymerase which initiate the replication of RNA genome of viruses in the host cell(s). For viruses containing single-stranded negative sense RNA (ss-RNA) genome, their ss-RNA genome must first be converted to a +RNA strand.
The +RNA strand is used as an mRNA template for transcription to the genomic –RNA genome. And this is enzymatically catalyzed by RNA-dependent RNA polymerase proteins. In the replication of double stranded RNA (dsRNA) genome of dsRNA viruses, the –RNA strand of the dsRNA genome must first be converted to a complementary +RNA strand that will serve as a template for the replication of the viral genome. RNA-dependent RNA polymerase also plays a role in the replication of the viral genome.
Retroviruses (e.g. human immunodeficiency virus, HIV) are RNA viruses that differ markedly from other RNA viruses in that the retroviruses synthesize mRNA and replicate their genome by means of an RNA-dependent DNA polymerase (also known as reverse transcriptase, RT). Hepadnaviruses are another group of animal viruses that utilizes RT in the replication of their genome apart from the viruses in the Retroviridae family (e.g. Retroviruses).
The Retroviruses have an ss+RNA. They do not use their ss+RNA genome as an mRNA template in the replication of their genome because it is not used as a messenger RNA or message. Rather, the RNA genome of Retroviruses is first transcribed into complementary DNA strands by RT in a genetic process known as reverse transcription (which is quite different from the central dogma of molecular biology – in which DNA is transcribed to RNA and then translated to protein).
Double stranded DNA (dsDNA) viruses depend solely on the cellular DNA replication of their host cell since the mRNA of dsDNA viruses is transcribed in the same manner akin to DNA replication of host cells. For single stranded DNA (ssDNA) viruses, their ssDNA is first converted to a dsDNA molecule that is transcribed to form mRNA. The host RNA polymerase plays a role in the transcription of the genome of DNA viruses into mRNA; and the viral mRNAs are later translated to specific proteins in the ribosomes located within the cytoplasm of the host cell.
The newly synthesized viral proteins are transported back to the nucleus of the host cell where the progeny viral particles are assembled and released from the infected host cell(s). It is however, noteworthy that new viral proteins are required for the replication of a viral genome (whether DNA or RNA genome) within an infected host cell; and the synthesis of these viral proteins are encoded by mRNAs transcribed from the genome of the infecting virus.
And as aforementioned, the viral genomic RNA can also serve as the mRNA (especially in RNA viruses), and in others, the viral genome can serve as a template for the synthesis of the mRNA – which is to be translated for viral protein synthesis. And some viruses such as the Retroviruses contain reverse transcriptase or RNA-dependent DNA polymerase that helps to carry out the transcriptional process in which the RNA genome of the virus is converted to ds DNA that act as the template for the synthesis of mRNA by normal cellular enzymes of the infected cells.
With the exception of Poxviruses and Hepadnaviruses, all animal DNA viruses replicate in the nucleus of their host cells; and the replication of viral genome varies from one virus to another depending on the genome the virus possess and its configuration i.e. whether it is double-stranded (ds) or single-stranded (ss) and whether it is positive sense (+) or negative sense (—). All RNA viruses replicate in the cytoplasm of the infected host cells with the exception of Orthomyxoviruses and Retroviruses (which can replicate in both the nucleus and cytoplasm).
ASSEMBLY STAGE OF VIRAL REPLICATION
The assembly of synthesized viral proteins and other associated molecules or particles is critical for viral pathogenesis in living cells. The viral proteins and viral genome must be packaged into a complete virion or viral particle before it can be released from the host cell. Virions carry out a self-assembling mechanism of the viral proteins and viral genomes within the infected host cell. The release of naked viruses form the host cell after packaging is quite different from the release of enveloped viral particles. Viral assembly is the last stage in the life cycle of viral infection; and it is accompanied by the release of the complete viral particles from the infected host cell.
Viral replication can occur in any of two ways: lytic cycle of viral replication and lysogenic cycle of viral replication (Figure 2). The lytic cycle is the normal process of viral reproduction involving penetration of the cell membrane, nucleic acid synthesis, and lysis of the host cell (Figure 2). It involves the reproduction of viruses using a host cell to manufacture more viruses; the viruses then burst out of the cell. Viruses overtake a living host cell and use the cellular machinery of the host cell to reproduce and form its own molecules – since viruses are not capable of independent or self-replication.
And one of the ways the infecting virus may choose to leave the affected host cell is by destroying the host cell. Viruses usually leave their host cell by cutting (lysing) their way out of the host cell. This is called the lytic cycle of a virus replication. In a lytic cycle, the virus reproduces thousands to millions of times in just a few hours and they produce many viral progeny during this time or process. This result to the weakening of the host cell wall enough that the cell will lyse, or burst open, setting the army of new viruses free. The lytic cycle results in the destruction of the infected cell and its membrane unlike the lysogenic cycle which does not lead to the destruction of the host cell and its membrane.
The lysogenic cycle involves the incorporation of the viral genome into the host cell genome, this infecting the host cell from within (Figure 2). It is a form of viral reproduction involving the fusion of the nucleic acid of a bacteriophage with that of a host, followed by proliferation of the resulting prophage. In the lysogenic cycle, the phage DNA first integrates into the bacterial chromosome to produce the prophage. When the bacterium reproduces, the prophage is also copied and is present in each of the daughter cells. The daughter cells can continue to replicate with the prophage present or the prophage can exit the bacterial chromosome to initiate the lytic cycle. Prophage is the latent form of the virus genome that remains within the host cell without destroying it.
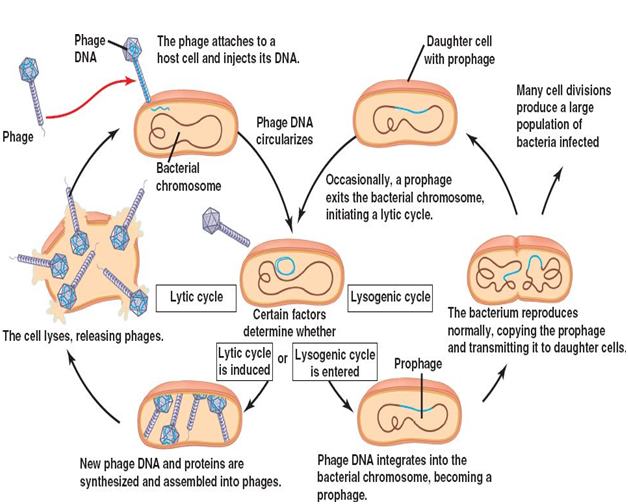
The difference between the lytic and lysogenic cycles of viral replication is that in the lytic phage, the viral DNA exists as a separate molecule within the bacterial (host) cell, and replicates separately from the host (bacterial) DNA while the location of viral DNA in the lysogenic cycle is within the host (bacterial) DNA. And while the lytic cycle of viral replication leads to the destruction of the host cell and membrane, the lysogenic cycle of viral replication does not lead to the destruction or lysis of the host cell and its membrane.
Lytic cycle is a type of viral life cycle that ends with host cell lysis and the release of numerous and newly synthesized viral progenies. This type of viral replication (i.e. lytic cycle) is usually carried out by virulent viruses that naturally lyse their host cells during the reproductive cycle. However, in lysogenic cycles, viruses engage in a different type of relationship with their host. And in this type of viral replication (i.e. lysogenic cycle), the viral genome does not take total control of the host and destroy it while it is still synthesizing new virions or phages as the case may be.
In lysogenic cycle, the viral genome remains within the host cell and replicates its own genome alongside the host (bacterial) genome to produce new cells that continue to grow and divide over a long period of time. The infected host cell looks normal; and each of the infected host cells can go on to produce new virions or phages which will lyse under normal conditions. The infecting virus establishes a type of relationship known as lysogeny – in which both the virus and the host cell coexist without destroying each other.
Lysogeny is defined as a relationship in which a virus or phage genome remains within its host cell (e.g. a bacterial cell) after infection and reproduces alongside the host genome instead of taking total control of the host cell and destroying it in the process of replication. The viruses that enter into this type of relationship with their host cell are known as temperate phages while the bacterial host cell that are able to produce new viral or phage particles under these conditions are known as lysogens; and they are said to be lysogenic.
RELEASE OF MATURE AND COMPLETE VIRIONS
Completely assembled and packaged viral genome and viral proteins are released from infected host cell as the self-assembly of the individual particles are complete. Naked viruses are usually released from the infected host cell via the lysis of host cells while enveloped viruses are released via the plasma or cell membrane of the host cell in a budding process in which the virion acquires its envelope in the process.
The morphogenesis of viral particles usually occurs simultaneously with viral release even though the process may vary in both naked viruses and enveloped viruses. While enveloped viruses mature by a budding process before their release from host cells as aforementioned, matured naked viruses do not undergo budding before they are released. Instead, mature naked viruses are immediately released from their host cells via apoptosis or cell lysis once the complete virion have been well assembled and packaged.
Upon the infection of new host cells, the virion disassembles spontaneously and penetration of the virus is initiated in the process of the viral infection. Virus-host interaction is critical to the initiation of a particular disease process in a host; and disease development is dependent on many factors including the amount of released viral particles; the host immune state; the type or number of organs infected by the virus; the virulence factors of the infecting virus; environmental factors and the persistence of the infecting virus within the host amongst other host and viral factors.
HOST IMMUNITY TO VIRAL INFECTION
Most viral infections do not result to disease development with the exception of AIDS and some haemorrhagic viral infections such as Ebola and Lassa fever that leads to serious disease in the affected host and even death. The effectiveness of the immune system of the viral-infected host is one singular factor there is that can significantly affect the outcome of a particular viral infection in an individual. Even though the immune system may not eliminate totally the infecting virus, a strong immune system helps to contain the pathogenicity or virulence of a given pathogenic virus.
The administration of potent vaccines and other antiviral agents to affected hosts and other susceptible host can help to treat the disease or infection and even prevent the contamination of a particular viral infection respectively. Vaccine development (which is mainly based on the isolation or cultivation, attenuation or inactivation and the direct or indirect injection of a susceptible host with killed, live or purified subunits of causative pathogenic microorganism) has saved mankind from the onslaught of some life-threatening infectious diseases such as measles and smallpox through the control and the complete eradication of these diseases in some cases.
The timely vaccination of a large population of susceptible host against a particular viral infection helped to minimize the spread of the pathogen in the immunized populace via herd immunity. And vaccination/immunization has helped to increase the life expectancy of mankind through the protection of the population from life-threatening diseases especially those caused by pathogenic viruses. Viral infection unlike other microbial infections stimulate an immune response in the host; and this serves to protect the host from immediate attack and even from futuristic viral infection through specific immune responses.
Pathogenic viruses as obligate intracellular microorganisms engage in uniquely intimate host-parasite relationships with the living organisms (plant, animal or man) that they infect, and this is due in part to the fact that viruses only exist or replicate in living cells. In the course of their pathogenicity or virulence in a particular host, pathogenic viruses express gene products that act to circumvent one or more of the several antiviral defense mechanisms (e.g. production of interferons) developed by the host organisms.
Nevertheless, the host resistance to viral infections involves both the humoral immunity and cell-mediated immunity. While some viral infections (e.g. influenza and common colds) can be contained by the host’s immune system; some others such as the causative agent of AIDS (i.e. HIV) overpowers the host’s immune system and makes it incapable to fight against the invading viral agent. Infections with some pathogenic viruses may lead to apoptosis; and this programmed cell death is a host defense mechanism that can be inhibited by some viruses.
Antibodies produced by humoral immunity can neutralize pathogenic viruses by interfering with their attachment to host cells; and the production of antibodies also enhances the destruction of viral particles via phagocytosis. However, antibodies cannot completely eliminate the infecting virus once the virion has incorporated its genome into that of the host. The cell-mediated immunity is one of the major important arms of the immune system that interfere with viral replication in vivo.
Activated lymphocytes including cytotoxic T lymphocytes (CTL), helper T cells (CD+4) and cytotoxic T cells (CD+8) can recognize and destroy viral infected cells especially when these organisms bud off from their host cells. The production of interferons (which are protein substances produced by cells during viral infection) helps to reduce the spread of virus especially in some benign viral infections such as influenzae and cold.
They stimulate the production of natural killer (NK) cells and T cells; and interferons also accelerate the immune response of a host to viral infection. And by acting on other effector cells of the immune system, interferons (which are antiviral cytokines) generally reduce the susceptibility of other uninfected cells of the host to the invading virus, and they do so by localizing the pathogenic virus so that they do not easily spread in the host’s body.
References
Acheson N.H (2011). Fundamentals of Molecular Virology. Second edition. John Wiley and Sons Limited, West Sussex, United Kingdom.
Alan J. Cann (2005). Principles of Molecular Virology. 4th edition. Elsevier Academic Press, Burlington, MA, USA.
Alberts B, Bray D, Johnson A, Lewis J, Raff M, Roberts K and Walter P (1998). Essential Cell Biology: An Introduction to the Molecular Biology of the Cell. Third edition. Garland Publishing Inc., New York.
Barrett J.T (1998). Microbiology and Immunology Concepts. Philadelphia, PA: Lippincott-Raven Publishers. USA.
Black, J.G. (2008). Microbiology: Principles and Explorations (7th ed.). Hoboken, NJ: J. Wiley & Sons.
Brian W.J Mahy and Mark H.C van Regenmortel (2010). Desk Encyclopedia of Human and Medical Virology. Elsevier Academic Press, San Diego, USA.
Brooks G.F., Butel J.S and Morse S.A (2004). Medical Microbiology, 23rd edition. McGraw Hill Publishers. USA.
Cann A.J (2011). Principles of Molecular Virology. Fifth edition. Academic Press, San Diego, United States.
Carter J and Saunders V (2013). Virology: Principles and Applications. Second edition. Wiley-Blackwell, New Jersey, United States.
Champoux J.J, Neidhardt F.C, Drew W.L and Plorde J.J (2004). Sherris Medical Microbiology: An Introduction to Infectious Diseases. 4th edition. McGraw Hill Companies Inc, USA.
Dimmock N (2015). Introduction to Modern Virology. Seventh edition. Wiley-Blackwell, New Jersey, United States.
Dimmock N.J, Easton A.J and Leppard K.N (2001). Introduction to modern virology. 5th edition. Blackwell Science publishers. Oxford, UK.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.