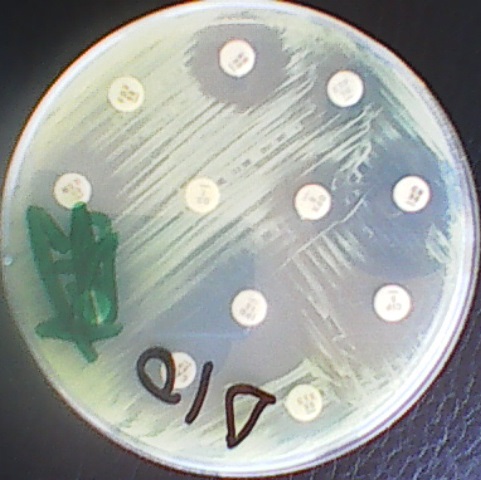
The introduction of antibiotics into clinical medicine for the treatment of infectious diseases heralded an era of one of the most significant breakthrough in medicine – since most infectious diseases that caused morbidity and mortality in the human race was drastically reduced and contained with antibiotics (the magic bullets). Particularly, penicillin was the first antibiotic (sourced from microbes) to be used clinically for the treatment of bacterial diseases; and this antibiotic proved very efficacious for the treatment of several bacterial related diseases.
Nevertheless, bacteria have now developed ingenious ways of evading the potent antibacterial and/or antimicrobial activities of some available antimicrobials including the penicillins; and this phenomenon makes some previously and easily treatable bacterial diseases to be difficult to treat now. The use of antimicrobial agents is believed to be the major driving force for the emergence and spread of microbial resistance; and the irrational use of drugs especially in animal husbandry, fish production, animal food technology and/or production, veterinary medicine and for other agricultural practices outside human medicine has significantly contributed to this dilemma.
Each time an antimicrobial or antibiotic is used (either rationally or irrationally); bacteria inevitably develop resistance mechanisms either through spontaneous mutations in the bacteria or through the acquisition of resistance genes from other resistant bacteria in its environment (acquired resistance). Acquired resistance is perhaps the most significant means through which bacteria acquire resistance genes and become resistant to an array of antimicrobials. And the horizontal transfer of genetic elements (especially resistance genes and/or plasmids) between bacteria in any environment is fundamental to the dissemination of resistance genes within a particular bacterial population in the presence of antibiotics.
The virulence determinants of bacteria aside their antibiotic resistance genes could easily be transferred within bacterial population under the selective pressure imposed by the presence of antimicrobial agents including antibiotics, disinfectants or a heavy metal. Bacteria can acquire resistance genes through several means of genetic transfer mechanisms including but not limited to conjugation (which involves direct cell to cell contact and transfer of transposons or plasmids); transduction (which is mediated by bacteriophages); and through transformation (which involves the uptake of free DNA that results from bacterial lysis).
There are different classes of antimicrobial agents or antibiotics used for the containment or control of microbes; and multidrug resistant microorganisms have developed several ways to resist the antimicrobial action of each and every one of these drug classes. Thus, a single bacterium can carry resistance genes to a totally unrelated series of antimicrobial agents. For example, a single pathogenic bacterium that is multidrug resistant in nature can carry antibiotic resistant genes that make the organism to be resistant to antibiotics in the beta-lactam class, aminoglycosides, fluoroquinolones and the macrolides.
Such an organism is called a multidrug resistant bacterium – since it carry genes that gives the organism an exceptional ability to be resistant to the antimicrobial action and/or onslaught of antibiotics in several drug classes including those in the beta-lactam group, aminoglycosides, macrolides and the aminoglycosides.
Multidrug resistant bacteria (MDRB) are bacterial strains that are resistant to a wide variety of antibiotics inclusive of the beta-lactams, the fluoroquinolones, macrolides, aminoglycosides and sulphamethoxazole-trimethoprim. They are bacteria that have become resistant to certain commonly used antibiotics; and these organisms are commonly found in different types of environment both in the hospital and in the community. MDRB are bacteria that have become resistant to more than one antibiotic. They are responsible for many hospital-acquired and community-acquired infections in humans.
MDRB can be acquired in several ways. They can be acquired through person to person contact or by sharing certain personal items with already infected or colonized persons. MDRB can also be acquired through contact with infected animals and also through contact with contaminated surfaces and fomites. People who are mostly at risk of infection with a multidrug resistant organism are those who are already ill and hospitalized, and people with weakened immune system.
The close proximity of sick patients in hospitals, receiving antimicrobial agents and who are often cared for by the same physician or healthcare personnel is also another contributing factor that could increase the chances of an individual to develop infections or diseases caused by a drug-resistant pathogen. People requiring frequent use of antibiotics such as cancer and transplant patients are also at risk of developing an infection caused by MDRB. Infections or diseases caused by multidrug resistant organisms can range from milder microbial infections to serious infections or diseases that can lead to even death.
MDRB can cause protracted urinary tract infections (UTIs), otitis media, pneumonia, skin and soft tissue infections, pelvic inflammatory disease (PID), gastroenteritis, diarrhea, endocarditis, bacteremia, septicaemia, respiratory tract infections and wound infections. The development, spread and persistence of multidrug resistant bacteria (MDRB) in both the community and hospital environment poses an increasing healthcare challenge to the public health; and this development threatens our ability to effectively threat some microbial diseases.
Some infectious agents and/or pathogenic microorganisms are now becoming increasingly and extensively resistant to some commonly available potent antimicrobial agents particularly antibiotics; and this phenomenon is one of our most public health threats owing to the morbidity and mortality associated with drug-resistant microbial infections. The loss of antimicrobial onslaught and efficacy will go a long way in undermining our ability to effectively fight and contain the nefarious activities of pathogens especially those that are multidrug resistant.
The treatment of MDRB is often difficult due to the resistance of the organisms to a wide variety of antibiotics. However, treatment of MDRB is often done using a combination of antibiotics and especially those antibiotics which are less commonly used. Of utmost importance to the treatment of infections caused by MDRB is proper antimicrobial susceptibility studies which will help guide the physician and/or the healthcare personnel on the best choice of antibiotics or drugs to use for a particular case. The emergence and spread of multidrug resistant strains of bacteria calls for an immediate action to combat and contain the situation, since this development defiles the antibiotic potency of some available antimicrobials.
Therapeutic failures associated to antimicrobial resistance increases the morbidity and mortality rates of infectious diseases; and this development impacts negatively on the economy of any people, their social status and the wellbeing of the affected individuals. In the face of antimicrobial resistance especially MDRB infections, the choice of drugs to be used for treatment are often limited; and such scenarios leads to severe clinical prognosis and potential treatment failures. It is therefore important to step up the search for newer antimicrobials that will contain the antibiotic-degrading potentials of MDRB.
And available antimicrobials should be used cautiously and only under a doctor’s prescription in order to preserve and protect the potency of these drugs. The use of antimicrobials especially those meant for human medicine in agricultural practices, food production, veterinary medicine, animal husbandry and in fish production should be discouraged – since these practices contribute a great deal to the emergence and spread of antibiotic resistant strains of bacteria in the community.
Typical examples of multidrug resistant bacteria include:
- Community-acquired methicillin resistant Staphylococcus aureus (CA-MRSA)
- Vancomycin resistant Enterococci (VRE)
- Vancomycin resistant Staphylococcus aureus (VRSA)
- Vancomycin intermediate Staphylococcus aureus (VISA)
- Klebsiella pneumoniae carbapenemase (KPC)
- Penicillin-resistant Streptococcus pneumoniae
- New Delhi metallo beta-lactamase (NDM-1)
- Carbapenem resistant Enterobacteriaceae (CRE)
- Multidrug resistant Acinetobacter baumannii (MDRAB)
- Extensively drug resistant tuberculosis (XDR-TB)
- Multidrug resistant Neisseria gonorrhoeae
- Multidrug resistant Pseudomonas aeruginosa
- Drug resistant non-typhoidal Salmonella
- Fluconazole-resistant Candida
- Drug-resistant Shigella
- Drug-resistant Salmonella Typhi
References
Arora D.R (2004). Quality assurance in microbiology. Indian J Med Microbiol, 22:81-86.
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Axelsen P.H (2002). Essentials of antimicrobial pharmacology. Humana Press, Totowa, New Jersey, USA. Al-Jasser A.M (2006). Extended – Spectrum Beta – Lactamases (ESBLs): A Global Problem. Kuwait Medical Journal, 38(3):171-185.
Bisht R., Katiyar A., Singh R and Mittal P (2009). Antibiotic Resistance – A Global Issue of Concern. Asian Journal of Pharmaceutical and Clinical Research, 2 (2):34-39.
Block S.S (2001). Disinfection, sterilization and preservation. 5th edition. Lippincott Williams & Wilkins, Philadelphia and London.
Cars O and Nordberg P (2005). Antibiotic resistance: The faceless threat. International Journal of Risk & Safety in Medicine, 17 (3/4): 103-110.
Carson C.F., Hammer K.A and Riley T.V (2006). Malaleuca alternifolia (Tea Tree) oil: A Review of Antimicrobial and other Medicinal Properties. Clinical Microbiology Review, 19(1):50-62.
Cowan M.M (1999). Plant products as antimicrobial agents. Clinical Microbiology Reviews., 564-582.
Denyer S.P., Hodges N.A and Gorman S.P (2004). Pharmaceutical Microbiology. 7th ed. Blackwell Publishing Company, USA.
Ejikeugwu Chika, Ikegbunam Moses, Ugwu Chigozie, Eze Peter, Iroha Ifeanyichukwu, and Esimone Charles (2013). Phenotypic Detection of Klebsiella pneumoniae Strains – Producing Extended Spectrum β-Lactamase (ESBL) Enzymes. Scholars Academic Journal of Biosciences. 1(1):20-23.
Ejikeugwu Chika, Iroha Ifeanyichukwu, Adikwu Michael and Esimone Charles (2013). Susceptibility and Detection of Extended Spectrum β-Lactamase Enzymes from Otitis Media Pathogens. American Journal of Infectious Diseases. 9(1):24-29.
Ejikeugwu Chika, Iroha Ifeanyichukwu, Adikwu Michael and Esimone Charles (2013). Susceptibility and Detection of Extended Spectrum β-Lactamase Enzymes from Otitis Media Pathogens. American Journal of Infectious Diseases. 9(1):24-29.
Finch R.G, Greenwood D, Norrby R and Whitley R (2002). Antibiotic and chemotherapy, 8th edition. Churchill Livingstone, London and Edinburg.
Joslyn, L. J. (2000). Sterilization by Heat. In S. S. Block (Ed.), Disinfection, Sterilization, and Preservation (5th ed., pp. 695-728). Philadelphia, USA: Lippincott Williams and Wilkins.
Lai P.K and Roy J (2004). Antimicrobial and chemopreventive properties of herbs and spices. Curr. Med. Chem, 11 (11): 1451–1460.
Livermore D.M (2004). The need for new antibiotics. Clinical Microbiology & Infection, 4(10): 1-9.
Mascaretti O.A (2003). Bacteria versus antibacterial agents: An integrated approach. Washington: ASM Press.
Nally J.D (Ed.) (2007). Good manufacturing practices for pharmaceuticals. Sixth edition. Informa Healthcare USA, Inc, New York.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.