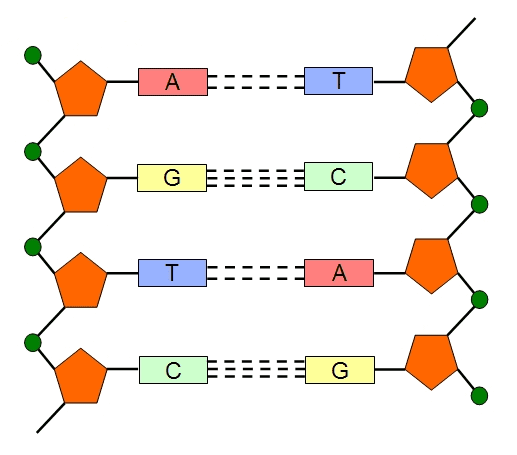
DNA replication is simply defined as the genetic process of duplicating the genome of a cell. It is an important process that goes on in the cell of every organism each time the organism undergoes reproduction; and DNA replication is a cellular process that occurs in a number of steps. DNA replication generally coordinates the process of cell division in living organisms inclusive of eukaryotic and prokaryotic cells. When an organism replicates its genome, it undergoes a duplication process which allows it to generate identical DNA molecules that is akin to the parent DNA.
This ensures that the daughter cells produced during cell division carry the same genetic information as the parent cell especially in mitotic cell division. DNA replication is a complex molecular process that occurs in cells, and it operates in a fast manner. Errors are minimized as the organisms DNA is being replicated. But when mutation occurs in the gene sequences being reproduced, the DNA replication process corrects it as the gene sequence is copied. It is noteworthy that genetic information always flows from the DNA to RNA which is finally translated in the ribosomes to form specific protein molecules as encoded by the organism’s gene or DNA.
It is noteworthy that the replication of bacterial chromosome normally starts at a fixed point generally known as the origin of replication (Ori); and replication proceeds in both directions of the cells chromosome (which is usually circular in shape) until it reaches a termination point (ter) where replication stops. Ori is the specific position on the DNA molecule of a cell where DNA replication begins. The termination point is approximately opposite to the origin of replication of the bacterial circular chromosome. The chromosome of some bacteria and eukaryotic cells is often linear in form; and DNA replication in these organisms is usually carried out in the same patterns with some variations.
The double stranded DNA separates into two individual strands during DNA replication; and each of the separated stands acts as a template for the synthesis of a new double stranded DNA molecule (Figure 1). DNA replication is a complex cellular reaction that is coordinated by several enzymatic and metabolic reactions within the cell of an organism. DNA replication can occur in vivo (i.e. inside a living cell such as E. coli in a natural manner) and in vitro (e.g. in PCR tubes). The circular double stranded DNA molecule is unwound or separated in vivo into two single strands by the activity of the enzyme, helicase – which unzips the double stranded DNA molecule into single strands (Figure 1).
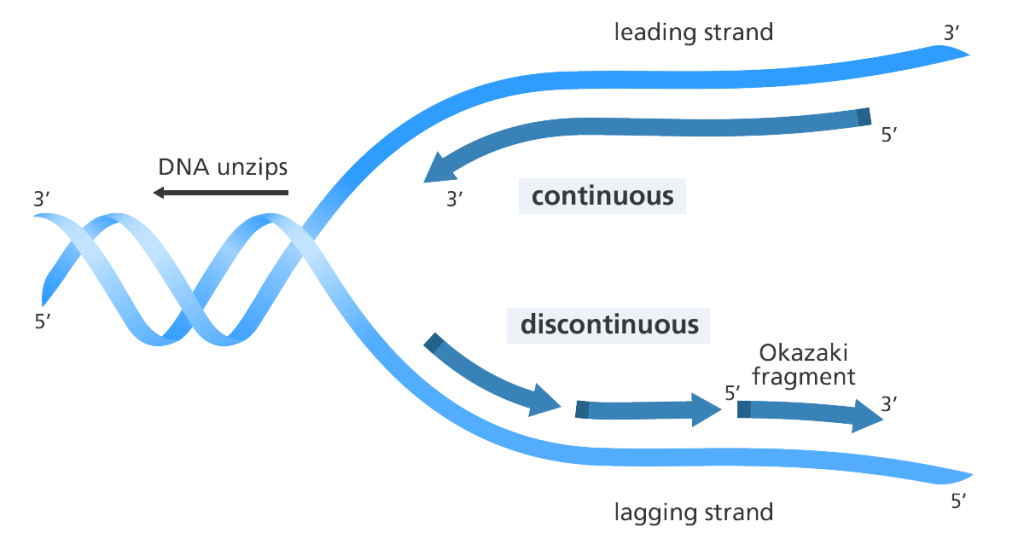
Helicase enzyme is important for DNA replication because without its action, DNA replication will not proceed. The enzyme binds to the double stranded DNA molecule and move along it to separate the DNA into two individual strands (i.e. single stranded DNA molecules). Once separated, each of the single strands is prevented from rejoining by the activities of special cellular proteins known as the single-stranded DNA binding proteins (SSBs). SSBs ensure that the single strands of DNA are kept separated after separation. The unwinding of the double helix of the DNA result in the formation of a Y-shaped structure known as the replication fork as shown in Figure 1.
It is noteworthy that each of the separated DNA single strands conserves and carries the genetic information encoded in the original double stranded parent DNA, and they also act as templates to direct the sequence of genetic information in the new strands. After unwinding, the two strands generated are known as the leading strand and the lagging strand respectively. Thereplication of the leading strand proceeds continuously because it possesses an exposed 3′ end to which nucleotides (e.g. dNTPs) are easily added in a complementary manner; and DNA replication in the leading strand or new daughter strand proceeds in the 5′-3′ direction.
DNA polymerase enzyme (e.g. DNA polymerase III) directs the synthesis of the leading strand in a continuous manner by the formation of phosphodiester bonds between the newly synthesized double stranded DNA. A short RNA primer (formed by RNA polymerase or primase) that is complementary to the parent DNA strand is required for the successful activity of DNA polymerase III in extended the DNA strand in a continuous fashion.
DNA synthesis or replication can only occur in the 5′-3′ direction, but this is not the case for the other strand of the replication fork known as the lagging strand. Replication of nucleic acid molecules in the 3′-5′ direction is not possible. The replication of the lagging strand proceeds in a discontinuous manner and in a 3′-5′ direction in short fragments known as the Okazaki fragments. These short fragments of DNA (i.e. the Okazaki fragments) are joined together to form a complete DNA strand by the activities of DNA polymerase I and DNA ligase enzymes.
The DNA ligase enzymes forms phosphodiester bonds between the Okazaki fragments after the RNA primers have been removed or preplaced by the DNA polymerase I enzyme. In the Okazaki fragments, nucleic acid synthesis is started with short pieces of RNA (which are produced by RNA polymerase enzyme). The RNA primer is extended by the action of DNA polymerase III, but it is later removed and the gap produced in the lagging strand is replaced by DNA polymerase I enzyme which ensures the extension of DNA molecule. DNA polymerase I enzyme exhibit a 5′-3′ exonuclease activity, and this enables it to remove RNA primer from the Okazaki fragments in the lagging strands, and replace same with complementary DNA.
And the fragments are joined together by DNA ligase enzymes to from new daughter duplexes. It is worthy of note that the newly synthesized daughter DNA strand is a composite of one parent strand and one newly synthesized strand. And this type of DNA replication in which the newly synthesized daughter duplexes consists of one parent strand and one newly synthesized strand is generally known as semiconservative replication because only one strand in each of the daughter duplex has been newly synthesized.
The semiconservative replication of DNA implies that half of the double stranded DNA molecules synthesized remain unchanged in both strands while the remaining half is changed. The termination of DNA synthesis occurs when the entire DNA duplex has been completely replicated. The circular DNA of microbial cells (especially the prokaryotes) has termination points on their DNA, and once the DNA polymerase reaches this site on the DNA, DNA replication ceases. Large amount of mutation during DNA replication, though rare will be deadly to the organism.
However, errors or mutation arising from DNA replication processes are contained by the DNA repair mechanisms of the cell. It is noteworthy that DNA is different from RNA even though that both are classified as nucleic acids (Table 1). Genetic information always flows from the DNA to the RNA which is later translated into protein molecules that perform various metabolic and cellular functions within the cell. The cellular functions of an organism are mainly performed by proteins whose capabilities are primarily determined by their genetic sequences as encoded in the DNA.
Table 1. DIFFERENCES BETWEEN DNA AND RNA
| DNA | RNA |
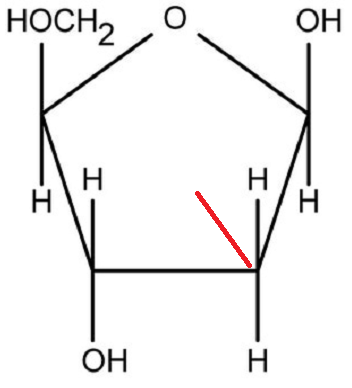
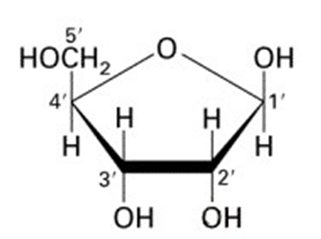
| DNA contains deoxyribose sugar (Figure 2). | RNA contains ribose sugar (Figure 3). |
| DNA does not have a fully hydroxyl group (OH–) at carbon number two (2) in its pentose sugar; and thus it lacks oxygen at this region. | RNA has a full hydroxyl group at carbon number two; and thus does not lack oxygen at this site. |
| DNA has thymine (T) as its pyrimidine or base. | RNA has uracil (U) as its pyrimidine or base. |
| DNA is a double stranded (ds) nucleic acid molecule. | RNA is a single stranded (ss) nucleic acid molecule. |
| DNA undergoes transcription to form mRNA. | RNA undergoes translation to form proteins. |
| DNA is primarily located in the nucleus of a cell. | RNA is located in the cytoplasm of a cell particularly within the ribosome where it directs protein synthesis. |
| DNA is a larger informational macromolecule than RNA | RNA is smaller than DNA. |
| DNA is the main genetic material of the cell because it contains the genetic sequence or genes that direct protein synthesis in the cell | RNA is not the main genetic material of the cell; and unlike DNA, the constituents of RNA inclusive of mRNA, rRNA and tRNA are chiefly responsible for the synthesis of proteins in the cell. |
References
Alberts B, Bray D, Johnson A, Lewis J, Raff M, Roberts K andWalter P (1998). Essential Cell Biology: An Introduction to the Molecular Biology of the Cell. Third edition. Garland Publishing Inc., New York.
Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., eds (2002). Short Protocols in Molecular Biology, 5th edn. John Wiley & Sons, New York.
Bains W (1998). Biotechnology: From A to Z. 2nd ed. Oxford University Press, New York, USA.
Branden C and Tooze J (1998). Introduction to protein structure. A non-technical introduction to protein structure. New York: Garland Press.
Chen I and Dubnau D (2004). DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2 (3): 241–249.
Cooper G.M and Hausman R.E (2004). The cell: A Molecular Approach. Third edition. ASM Press.
Dale J (2003). Molecular genetics of bacteria. Jeremy W. Dale and Simon Park (4th eds.). John Wiley & Sons Ltd, West Sussex, UK. Pp. 312-313.
Dale J (2003). Molecular genetics of bacteria. Jeremy W. Dale and Simon Park (4th eds.). John Wiley & Sons Ltd, West Sussex, UK. Pp.
Glick B.R and Pasternak J.J (2003). Molecular Biotechnology: Principles and Applications of Recombinant DNA. ASM Press, Washington DC, USA.
Hames B.D and Rickwood D (1998). Gel Electrophoresis of Proteins: A Practical Approach 3rd Edition. The Practical Approach Series, Oxford University Press
Latha C.D.S and Rao D.B (2007). Microbial Biotechnology. First edition. Discovery Publishing House (DPH), Darya Ganj, New Delhi, India.
Lewis R (2007). Human Genetics: Concepts and Applications. Seventh edition. McGraw-Hill Companies, Inc, New York, USA.
Lodish H, Berk A, Matsudaira P, Kaiser C.A, Kreiger M, Scott M.P, Zipursky S.L and Darnell J (2004). Molecular Cell Biology. Fifth edition. Scientific American Books, Freeman, New York, USA.
Madigan M.T., Martinko J.M., Dunlap P.V and Clark D.P (2009). Brock Biology of Microorganisms, 12th edition. Pearson Benjamin Cummings Inc, USA.
Tamarin Robert H (2002). Principles of Genetics. Seventh edition. Tata McGraw-Hill Publishing Co Ltd, Delhi.
Thieman W.J, Palladamo M.A and Thieman W (2003). Introduction to Biotechnology. Benjamin Cummings, San Francisco, CA.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.