The expression “phenotypic” is from the word phenotype, which means “the observable characteristics of an organism”. It describes the physical features of an organism in terms of its morphology, motility, colour and shape amongst other noticeable attributes that are typical to a particular organism. Thus, the phenotypic detection and characterization of antimicrobial resistant genes in pathogenic bacteria describes the general physical reaction or observable traits of microbes to a variety of antimicrobial agents such as antibiotics, disinfectants and disinfectants. In phenotypic detection, pathogenic strains of microorganisms are directly tested in vitro for their susceptibility or resistance to a range of antimicrobial agents (especially antibiotics) that are usually used for their control.
The result obtained from a phenotypic detection and characterization experiment generally gives a picture of the performance of the test drugs in vivo (i.e. when used for the practical treatment of a sick host). Phenotypic detection methods provides prompt and reliable drug susceptibility results that give the microbiologist a picture of the prevailing resistant profile of the test pathogen, and this guides physicians on the choice of antimicrobial therapy to initiate in the patient. Phenotypic detection and characterization techniques used in the microbiology laboratory to classify resistant genes of pathogenic bacteria are easy to perform and report that their genotypic counterparts. Some phenotypic detection methods (e.g. the VITEK system) are automated and can be used to perform computerized or automated antimicrobial susceptibility testing in the microbiology laboratory.
MicroScan and Phoenix systems are other available automated antimicrobial susceptibility methods that can be used in the microbiology laboratory to promptly evaluate the susceptibility profile of pathogenic bacteria based on their MIC values. Phenotypic decoction and characterization techniques for the identification of resistance genes in pathogenic bacteria are generally less expensive and straightforward to perform. Most importantly, they can be used to monitor the progress and response of a diseased patient to antimicrobial therapy. One of the extensively used and generally accepted methods of characterizing the resistant genes of pathogenic bacteria by phenotypic methods in the microbiology laboratory is the Kirby-Bauer disk diffusion technique.
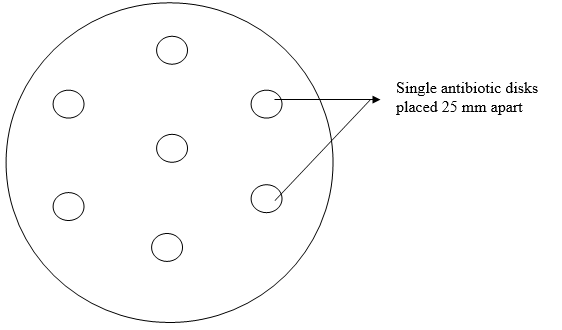
The Kirby-Bauer disk diffusion technique is one of the oldest approaches to the phenotypic antimicrobial susceptibility testing of pathogenic bacteria, and it still remains one of the most widely used antimicrobial susceptibility testing methods in the routine susceptibility studies of pathogenic microorganisms. It is most suitable for testing the majority of bacterial pathogens that are commonly encountered in the microbiology laboratory; and it is usually carried out using a template to avoid any form of bias when performing the test (Figure 1). Briefly, when a paper disk impregnated with a known concentration of a drug is placed on a agar plate, moisture from the agar plate is immediately absorbed into the drug laden disk. The drug in the disk begins to diffuse into the surrounding area of the agar which has been previously inoculated with the test bacteria and incubated at the appropriate temperature.
Bacterial growth occurs in the presence of an antibiotic when the bacteria reaches a critical mass and can overpower the inhibitory effects of the drug in the paper disk. The point at which critical mass of the bacteria is reached is demonstrated by a sharply marginated circle of bacterial growth around the paper disk (Figure 2).
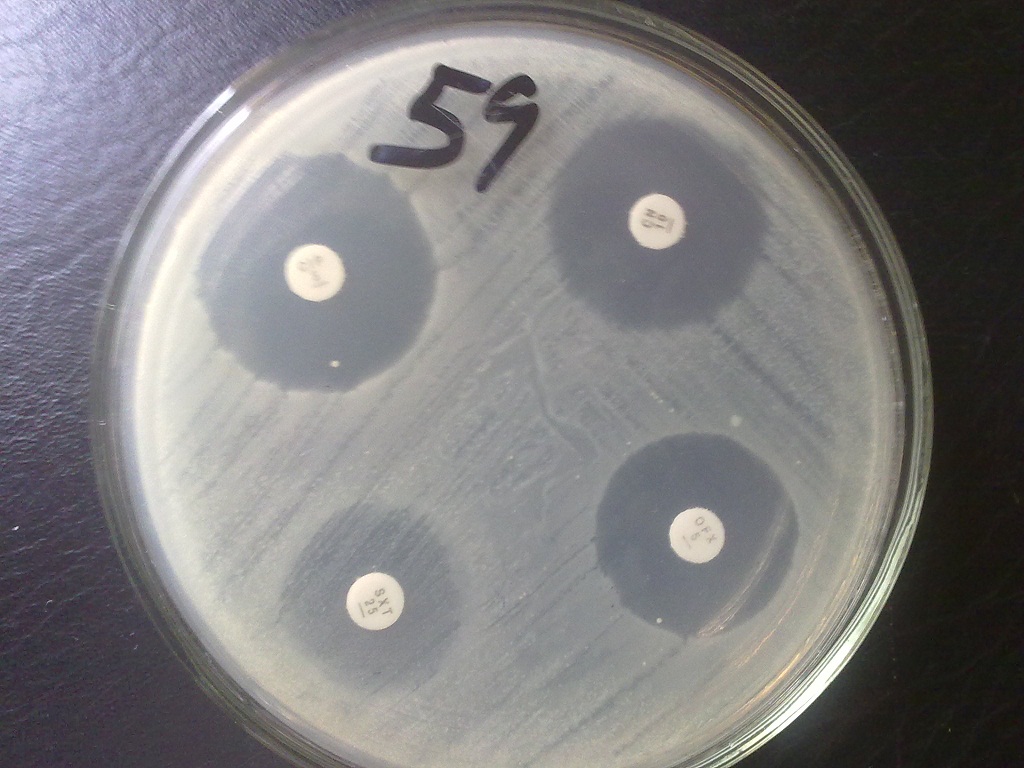
When the test organism is resistant to the antimicrobial agents used, a smaller zone of inhibition is usually produced (Figure 3). The marginated circle of bacterial growth produced around the antimicrobial disk is often referred to as the inhibition zone of the drug and it is measured to the nearest millimeter using a meter rule.
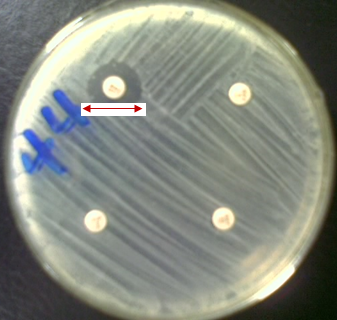
The zone of inhibition is usually compared to standard inhibition zone diameters (antibiotic breakpoints) as per the criteria of the Clinical Laboratory Standard Institute, CLSI (formerly known as the National Committee for Clinical Laboratory Standard, NCCLS. Observed inhibition zone diameters are normally reported as sensitive, intermediate or resistant in line with the CLSI criteria. Generally, the size of the IZD of the drug against the test pathogenic bacteria is used to evaluate the in vivo susceptibility of the organisms to the tested drug. In addition to the inhibition zone diameter (IZD) of a drug to pathogenic bacteria, the phenotypic decoction and characterization of antibiotic resistance gene also gives a clue of the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of the tested drug.
References
Arora D.R (2004). Quality assurance in microbiology. Indian J Med Microbiol, 22:81-86.
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Barenfanger J, Drakel C and Kacich (1999). Clinical and Financial Benefits of Rapid Bacterial Identification and Antimicrobial Susceptibility Testing. Journal of Clinical Microbiology, 37(5):1415-1418.
Denyer S.P., Hodges N.A and Gorman S.P (2004). Pharmaceutical Microbiology. 7th ed. Blackwell Publishing Company, USA.
Doern G, Brueggemannn A.B, Perla R, Daly D, Halkias D, Jones R.N, Saubolle M.A (1997). Multicenter laboratory evaluation of the bioMerieux Vitek antimicrobial susceptibility testing system with 11 antimicrobial agents versus members of the family Enterobacteriaceae and Pseudomonas aeruginosa. J Clin Microbiol, 35:2115–2119.
Doern G, Vautour R, Gaudet M, Levy B (1994). Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol, 32:1757–1762.
Doern G.V (1995). Susceptibility tests of fastidious bacteria. Manual of Clinical Microbiology, 6th edition, Murray P.R, Baron E.J, Pfaller M.A, Tenover F.C, Yolken R, American Society for Microbiology, Washington DC, Pp. 1342-1349.
Funke G, Monnet D, deBernardis C, von Graevenitz A, Freney J (1998). Evaluation of the VITEK 2 system for rapid identification of medically relevant Gram-negative rods. J Clin Microbiol, 36:1948–1952.
Garcia L.S (2010). Clinical Microbiology Procedures Handbook. Third edition. American Society of Microbiology Press, USA.
Hart C.A (1998). Antibiotic Resistance: an increasing problem? BMJ, 316:1255-1256.
Livermore D.M, Winstanley T.B, Shannon K.P (2001). Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes.
J Antimicrob Chemother, 48 Suppl 1:87-102.
Madigan M.T., Martinko J.M., Dunlap P.V and Clark D.P (2009). Brock Biology of Microorganisms, 12th edition. Pearson Benjamin Cummings Inc, USA.
Mahon C. R, Lehman D.C and Manuselis G (2011). Textbook of Diagnostic Microbiology. Fourth edition. Saunders Publishers, USA.
National Committee for Clinical Laboratory Standards. Performance Standards for antimicrobial susceptibility testing. 8th Informational Supplement. M100 S12. National Committee for Clinical Laboratory Standards, 2002. Villanova, Pa.
Washington J.A (1993). Rapid antimicrobial susceptibility testing: technical and clinical considerations. Clin Microbiol Newsl, 15:153–155.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.