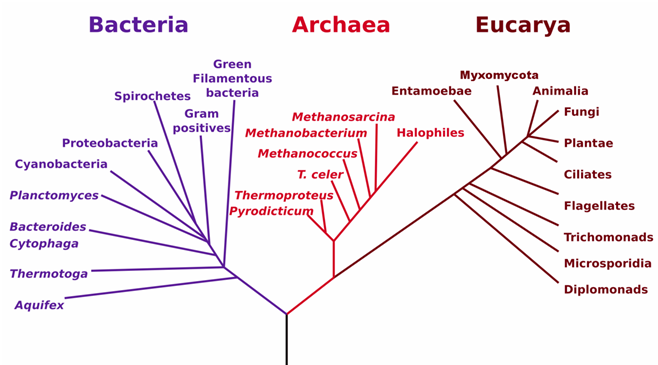
The phylogenetic tree of life comprises mainly of Eubacteria, Archaea and Bacteria. Eubacteria or Eukarya, Archaea and Bacteria are the three primary groups of microorganisms and macroorganisms, and these are generally referred to as domains- which show the ancestry of these organisms according to phylogeny. Phylogeny is the branch of science that studies the evolutionary correlation amongst organisms using the genetic information encoded in their nucleic acids (i.e. DNA and RNA). However, microbial phylogeny (i.e. the study of the evolutionary relationship between microbes) can be best deduced and studied by using the ribonucleic acid (RNA) component of an organisms nucleic acid and this is because all microbial cells contain ribosomes (the cell structure or component that is composed of RNAs) with the exception of viruses. Ribosomes are the machinery responsible for the synthesis of proteins in a cell; and they are found in all microbial cells except viruses.
Viruses contain either DNA or RNA unlike other microbial cells such as bacteria and fungi that contain both DNA and RNA in their genome. Thus, the RNA macromolecule especially the ribosomal RNA (rRNA) is the best tool for deciphering the evolutionary relationships of microbial cells excluding viruses. The phylogenetic relationship of viruses is determined is determined based on epidemiological, immunological, and evolutionary data or processes that are peculiar to viruses; and these information help biologists to form viral phylogenies – since they cannot be placed in the usual phylogenetic tree of life like other microbial cells. Viruses cannot be included in the phylogenetic tree of life as is applicable to other microbes such as bacteria because viruses do not share characteristics with other microbial cells, and no single gene is shared by all viruses or viral lineages. Thus, viruses are polyphyletic. It is noteworthy that in a phylogenetic tree, the characteristics of members of taxa are inherited from previous ancestors of organisms in those taxa (Figure 1).
Viruses are practically not alive like bacteria, fungi, protozoa and algae especially when they are outside a living host; and the generally lack any form of energy and carbon metabolism that keep living cells going. Viruses cannot replicate or evolve like other microbial cells; and they are reproduced only within living host cells including those of bacteria, plant and animal cells. Without these living host cells (which provide the reproductive machinery for the replication of viruses), viruses are generally inanimate complex organic matter. While cellular life has a single, common origin, viruses are polyphyletic – they have many evolutionary origins.
The phylogenetic tree which elucidates the three domains of life (Eukarya, Archaea and Bacteria) is constructed based on the application of the comparative sequencing of the rRNAs of different organisms (Figure 1). Eubacteria, Archaea and Bacteria are morphologically and genetically distinct organisms. The prokaryotes are only in two domains: Bacteria and Archaea while the eukaryotes are in the domain Eukarya. All organisms in these domains especially the prokaryotes have a wide range of physiological and genetic diversity that distinguishes them from each other. It is noteworthy that all microbial cells (bacteria, fungi, algae, protozoa and viruses) evolve with time; and thus newer species and diversities of organisms will continue to appear on the face of the earth due to natural selection and mutation and other prevailing environmental and non-environmental factors.
References
Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006). Toward automatic reconstruction of a highly resolved tree of life. Science, 311(5765):1283–1287.
Euzéby, J. P. (2010). List of Bacterial Names with Standing in Nomenclature. Int. J. Syst. Bacteriol., 47, 8 July, 2016. Retrieved from http://www.bacterio.cict.fr/m/micrococcus.html
Fauquet C.M, Mayo M.A, Maniloff J, Desselberger U and Ball L.A. (Eds) (2005). Virus Taxonomy. Eight Report of the International Committee on Taxonomy of Viruses. Burlington, M.A. Elsevier Academic Press.
George M. Garrity (2005). Bergey’s manual of systematic bacteriology. 2. Auflage. Springer, New York, 2005, Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria.
Grenfell B.T, Pybus O.G, Gog J.R, Wood J.L, Daly J.M, Mumford J.A and Holmes E.C (2004). Unifying the Epidemiological and Evolutionary Dynamics of Pathogens. Science, 303(5656): 327–332.
Gupta RS (2000). The natural evolutionary relationships among prokaryotes”. Crit. Rev. Microbiol, 26 (2):111–131.
Janda, J. M., and Abbott, S. L. (2006). The Genera Klebsiella and Raoultella. The Enterobacteria (2nd ed., pp. 115-129). Washington, USA: ASM Press.
Madigan M.T., Martinko J.M., Dunlap P.V and Clark D.P (2009). Brock Biology of microorganisms. 12th edition. Pearson Benjamin Cummings Publishers. USA. Pp.795-796.
Moreira D and López-García P (2009). Ten reasons to exclude viruses from the tree of life Nature Reviews Microbiology, 7(4):306-311.
Murray P.R, Baron E.J, Jorgensen J.H., Pfaller M.A and Yolken R.H (2003). Manual of Clinical Microbiology. 8th edition. Volume 1. American Society of Microbiology (ASM) Press, Washington, D.C, U.S.A.
Purves W.K, Sadava D, Orians G.H, and Heller H.C (2007). Life: The Science of Biology, 7e by Sinauer Associates, W.H. Freeman and Company, Sunderland.
Salyers A.A and Whitt D.D (2001). Microbiology: diversity, disease, and the environment. Fitzgerald Science Press Inc. Maryland, USA.
Woese C.R, Kandler O, Wheelis M.L (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Science, 87(12):4576–4579.
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.