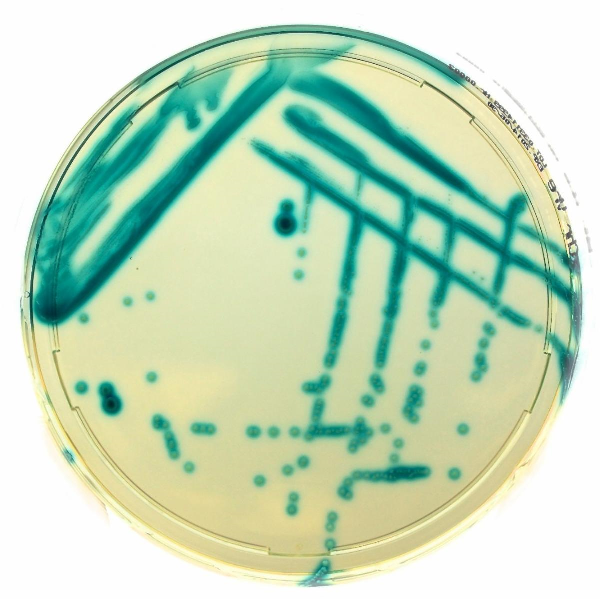
Chromatic VRE is a selective and differential chromogenic medium used for the qualitative and presumptive detection of vancomycin-resistant Enterococci directly from clinical samples. This medium is intended as an aid in the detection of the following bacteria: Enterococcus faecalis and Enterococcus faecium with acquired vancomycin resistance.
Chromatic VRE agar is not intended to diagnose infection or guide therapy. Results can be interpreted after incubation for 24 hours. Subculture to a non-selective medium is required for confirming identification, antimicrobial susceptibility testing and epidemiological typing.
Chromatic VRE agar contains a mixture of antibiotics including vancomycin for screening Vancomycin-resistant enterococci (VRE) and provides presumptive identification of Enterococcus faecium and Enterococcus faecalis directly from clinical specimens.
VRE have recently been recognized as one of the most severe cause of nosocomial infections.
An intrinsic resistance gene (vanC, vanD, vanE, vanF etc) is found in E. gallinarum and E. casseliflavus/E. flavescens and shows low resistance to vancomycin. Instead, an acquired resistance of vancomycin in enterococci (vanA & vanB types) is mostly detected in E. faecium and E. faecalis.
Constituents of Chromatic VRE agar (Figure 1)
Peptones 30.0g/L
Sodium Chloride 5.0g/L
Chromogenic and Selective Mix 6.5g/L
Agar 15.0g/L
Final pH 7.2 ± 0.2 at 25°C
The prompt detection of Vancomycin-resistance of E. faecium and E. faecalis is basic for avoiding the spread of this resistance to more virulent such as S. aureus.
Vancomycin Resistant Enterococci (VRE) was first isolated in Europe in the late 1980s and recently have been associated with nosocomial infections around the world. Acquisition of vancomycin resistance is mediated by several van gene clusters. The most commonly encountered vancomycin resistance genes are vanA and vanB, which are highly transferable between enterococci, mostly E. faecium and E. faecalis, and can be transferred to other organisms, including S. aureus (VRSA).
E. faecium is the most frequently isolated species of VRE in hospitals and typically produces high vancomycin (>128 µg/ml) and teicoplanin (≥16 µg/ml) minimum inhibitory concentrations (MICs). These isolates typically contain vanA genes. A vanB-containing isolate typically produces lower level resistance to vancomycin (MIC 16 to 64 µg/ml) and is susceptible to teicoplanin (MIC ≤1 µg/ml). In contrast to vanA and vanB, vanC and vanD in VRE are chromosomally associated, i.e. much less transferable.
The vanC gene generally presents with low-level resistance against vancomycin (MIC 2–32 µg/ml) and teicoplanin (MIC ≤1 µg/ml). Moreover, VRE containing vanC are typically E. gallinarum or E. casseliflavus/E. flavescens, and associated with low risk of mortality and rarely cause nosocomial outbreaks. The vanD gene, generally, confers moderate-level resistance against vancomycin (MIC 64–128 µg/ml) and teicoplanin (MIC 4– 64 µg/ml) and is constitutively expressed.
To limit the spread of glycopeptide resistance between enterococci and to more pathogenic bacteria which in turn can lead to clinical isolates resistant to all antibiotics, several public health institutes now screen for the carriage of vanA- and vanB-positive VRE in hospitalized patients.
Principle behind the Chromatic VRE agar
- Peptones provide amino acids, nitrogen, minerals, vitamins, and nutrient factors for growth of bacteria. Sodium chloride maintains the osmotic balance of the medium.
- The chromogenic and selective mix facilitates the identification of bacteria on the basis of the color and colony morphology, inhibiting most of yeasts and bacteria with the exception of VRE.
- Agar is the solidifying agent.
Source:
http://www.liofilchem.net/login/pd/ifu/11621_IFU.pdf
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.