ESBLs (extended spectrum beta-lactamases) are enzymes that mediate resistance to extended-spectrum (third generation) cephalosporins (e.g., ceftazidime, cefotaxime, and ceftriaxone) and monobactams (e.g., aztreonam) but do not affect cephamycins (e.g., cefoxitin and cefotetan) or carbapenems (e.g., meropenem or imipenem).
Why should hospital laboratories and hospitals be concerned about detecting ESBL-producing bacteria?
The presence of an ESBL-producing organism in a clinical infection can result in treatment failure if one of the above classes of drugs is used. ESBLs can be difficult to detect because they have different levels of activity against various cephalosporins. Thus, the choice of which antimicrobial agents to test is critical.
For example, one enzyme may actively hydrolyze ceftazidime, resulting in ceftazidime minimum inhibitory concentrations (MICs) of 256 µg/ml, but have poor activity on cefotaxime, producing MICs of only 4 µg/ml. If an ESBL is detected, all penicillins, cephalosporins, and aztreonam should be reported as resistant, even if in vitro test results indicate susceptibility.
How can the clinical microbiology laboratory screen for ESBL-producing bacteria?
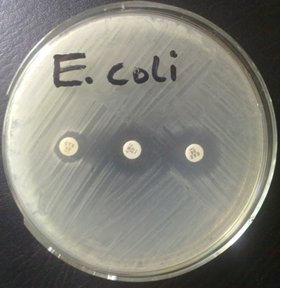
The Clinical Laboratory Standard Institute, CLSI (formerly: National Committee for Clinical Laboratory Standards, NCCLS) has developed broth microdilution and disk diffusion screening tests using selected antimicrobial agents. Each Klebsiella pneumoniae, K. oxytoca, or Escherichia coli isolate should be considered a potential ESBL-producer if the test results are as follows:
| Disk diffusion | MICs |
| cefpodoxime < 22 mm | cefpodoxime > 2 µg/ml |
| ceftazidime < 22 mm | ceftazidime > 2 µg/ml |
| aztreonam < 27 mm | aztreonam > 2 µg/ml |
| cefotaxime < 27 mm | cefotaxime > 2 µg/ml |
| ceftriaxone < 25 mm | ceftriaxone > 2 µg/ml |
The sensitivity of screening for ESBLs in enteric organisms can vary depending on which antimicrobial agents are tested. The use of more than one of the five antimicrobial agents suggested for screening will improve the sensitivity of detection. Cefpodoxime and ceftazidime show the highest sensitivity for ESBL detection.
How can clinical laboratory personnel confirm ESBL production?
CLSI recommends performing phenotypic confirmation of potential ESBL-producing isolates of K. pneumoniae, K. oxytoca, or E. coli by testing both cefotaxime and ceftazidime, alone and in combination with clavulanic acid. Testing can be performed by the broth microdilution method or by disk diffusion. For MIC testing, a decrease of > 3 doubling dilutions in an MIC for either cefotaxime or ceftazidime tested in combination with 4 µg/ml clavulanic acid, versus its MIC when tested alone, confirms an ESBL-producing organism. For disk diffusion testing, a > 5 mm increase in a zone diameter for either antimicrobial agent tested in combination with clavulanic acid versus its zone when tested alone confirms an ESBL-producing organism.
What quality control organisms should laboratories use for ESBL detection?
K. pneumoniae ATCC 700603 (positive control) and E. coli ATCC 25922 (negative control) should be used for quality control of ESBL tests.
Can an ESBL be present in an isolate of K. pneumoniae that is resistant to ceftazidime and/or cefotaxime, but demonstrates no clavulanic acid effect in the phenotypic confirmatory test?
Yes. The phenotypic confirmatory test does not detect all ESBLs. Some organisms with ESBLs contain other β-lactamases that can mask ESBL production in the phenotypic test, resulting in a false-negative test. These β-lactamases include AmpCs and inhibitor-resistant TEMs (IRTs).
Hyper-production of TEM and/or SHV β-lactamases in organisms with ESBLs also may cause false-negative phenotypic confirmatory test results. Currently, detection of organisms with multiple β-lactamases that may interfere with the phenotypic confirmatory test can only be accomplished using isoelectric focusing and DNA sequencing, methods that are not usually available in clinical laboratories.
How should cephalosporin and penicillin results be reported?
If an isolate is confirmed as an ESBL-producer by the CLSI-recommended phenotypic confirmatory test procedure, all penicillins, cephalosporins, and aztreonam should be reported as resistant.
This list does not include the cephamycins (cefotetan and cefoxitin), which should be reported according to their routine test results.
If an isolate is not confirmed as an ESBL-producer, current recommendations suggest reporting results as for routine testing. Do not change interpretations of penicillins, cephalosporins, and aztreonam for isolates not confirmed as ESBLs.
Do isolates other than K. pneumoniae, K. oxytoca, or E. coli produce ESBLs?
Yes. Other isolates of Enterobacteriaceae, such as Salmonella species and Proteus mirabilis, and isolates of Pseudomonas aeruginosa produce ESBLs. However, at this time, methods for screening and phenotypic confirmatory testing of these isolates have not been determined by CLSI.
Source:
CLSI
Discover more from Microbiology Class
Subscribe to get the latest posts sent to your email.