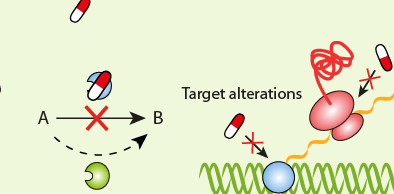
The resistance of a microbial cell to the potent action of antimicrobial agents or antibiotics can either be innate or acquired. In innate (natural) resistance for example, the microorganisms are naturally resistant to a particular antibiotic. This usually occurs in microbes that lack target sites for the binding of the antibiotic. But in acquired resistance, microorganisms acquire resistance genes from their surrounding environment or from other resistant organisms and incorporate the resistant trait or gene into their own genetic functions. This allows them to go on expressing resistance genes even as they proliferate and pass on the trait or gene to other susceptible microbes in their surroundings.
The successful treatment and management of an infectious diseases or infection caused by a resistant pathogen is usually based on a proper understanding of the correlation between the resistant genes produced by the invading-resistant pathogen and the activity of drugs directed towards it. Resistant strains of microorganisms are notorious in producing a wide variety of genes and/or extracellular enzymes which they use to neutralize the killing or inhibitory effect any antibiotics or drugs directed to them.
This scenario allows resistant microbes to continue to thrive and cause more harm to the patient even in the face of potent antimicrobial onslaught. Bacteria have evolved to survive in diverse environments. They survive exposure to harsh chemicals including antibiotics, and they also survive difficult growth conditions. They have learned to “detoxify” harmful substances e.g. antibiotics. Antibiotic resistance can either be intrinsic or acquired.
INTRINSIC (INNATE) RESISTANCE
Some bacteria are said to possess innate/intrinsic resistance against antibacterial action put forward by antibiotics. These microbes mount a great ingenuity in devising means or ways of neutralizing the killing or growth inhibitory action of antibiotics directed towards them. This innate form of antibiotic resistance in bacteria shows the different variations in the structure of the cell envelope of the organism, which allows them to mount resistance against drugs. The structural makeup of some bacteria also makes them to be naturally resistant to antibiotics.
For example, the cell wall of Gram negative bacteria contain outer membrane (OM) that covers the bacterial cell wall and thus makes it difficult for antibiotics to reach the cell wall of the organism. Innate (natural) resistance is a vertical means by which bacteria acquire resistance; and this usually occurs through spontaneous mutation. In vertical gene transfer (VGT), microbes transfer their genetic material or DNA to their progeny during cell division and/or DNA replication. Once a spontaneous mutation leading to the development of resistance genes occur in a bacterium; the resistance genes so developed can be passed on to all the bacteria’s progeny during cell division or DNA replication.
This type of mutation is rare in bacterial population. But when it occurs, the subsequent progeny acquire the resistance genes formed. A spontaneous mutation in the bacterial chromosome imparts resistance to a member of the same bacterial population, and this continues as the organism undergoes DNA replication. This exemplifies how genes or DNA can be transferred amongst microbial populations via a vertical gene transfer mechanism. Intrinsic or innate form of antibiotic resistance can occur by any one of the following route:
- Spontaneous mutation in the chromosomal DNA of bacteria.
- Accumulation of several point mutations in bacteria.
- An evolutionary process occurring only under selective pressure such as in the prior exposure of bacteria to antibiotics.
ACQUIRED (PHENOTYPIC) RESISTANCE
Microorganisms develop numerousmechanismsincluding mutation in order to acquire resistance to antimicrobial agents or antibiotics. Acquired resistance is a type of antibiotic resistance that is acquired by bacteria from their environment or from other drug resistant microorganisms by one of the mechanisms of genetic transfer. Conjugation, transformation, and transduction are typical examples of genetic transfer mechanisms via which microorganisms can transfer their genetic material (i.e. the DNA) from one organism to another.
Conjugation is a direct cell-to-cell contact between two bacteria via which the transfer of small pieces of DNA called plasmids takes place. Transformation is a process where parts of a DNA are taken up by the bacteria from the external environment. Transduction occurs when bacteriophages or phage (i.e. bacteria-specific viruses) transfer DNA between two closely related bacteria. In acquired/phenotypic resistance, the bacteria acquire reduced susceptibility to antibiotics through adaptation to growth within a specific environment (such as undue exposure to antibiotics).
Acquired resistance is a horizontal means by which bacteria become resistant to the antibacterial properties of antibiotics. In horizontal gene transfer (HGT), the genetic material or DNA of a given organism can be transferred between individual bacterial of the same species or even amongst individuals of different species. In acquired resistance, the organism acquires new genetic material (especially those that mediate antibiotic resistance) from another organism or from other sources. Acquired resistance can be achieved in bacteria by several routes as follows:
- Resistance can be maintained on horizontal mobile elements like plasmids, integrons and transposons. These mobile genetic elements serve as route through which antimicrobial or antibiotic resistance genes can be passed on from one organism to another through genetic transfer mechanisms such as conjugation.
- Resistant genes can be transferred among bacteria through one of the mechanisms of genetic transfer including transduction, conjugation and transformation.
- Resistance genes can be integrated into the bacterial chromosome or can be maintained in an extra chromosomal state (e.g. plasmids).
References
Arora D.R (2004). Quality assurance in microbiology. Indian J Med Microbiol, 22:81-86.
Ashutosh Kar (2008). Pharmaceutical Microbiology, 1st edition. New Age International Publishers: New Delhi, India.
Axelsen P.H (2002). Essentials of antimicrobial pharmacology. Humana Press, Totowa, New Jersey, USA. Al-Jasser A.M (2006). Extended – Spectrum Beta – Lactamases (ESBLs): A Global Problem. Kuwait Medical Journal, 38(3):171-185.
Ejikeugwu Chika, Ugwu Malachy, Iroha Ifeanyichukwu, Gugu Thaddeus, Duru Carissa, Eze Peter, and Esimone Charles (2013). Detection and antimicrobial susceptibility of some Gram negative bacteria producing carbapenemases and extended spectrum beta lactamases. International Journal of Microbiology and Immunology Research, 2(6):064-069.
Ejikeugwu P.C., Ugwu C.M., Araka C.O., Gugu T.H., Iroha I.R., Adikwu M.U and Esimone C.O (2012). Imipenem and Meropenem resistance amongst ESBL producing Escherichia coli and Klebsiella pneumoniae clinical isolates. International Research Journal of Microbiology. 3(10):339-344.
Finch R.G, Greenwood D, Norrby R and Whitley R (2002). Antibiotic and chemotherapy, 8th edition. Churchill Livingstone, London and Edinburg.
Joslyn, L. J. (2000). Sterilization by Heat. In S. S. Block (Ed.), Disinfection, Sterilization, and Preservation (5th ed., pp. 695-728). Philadelphia, USA: Lippincott Williams and Wilkins.
Lai P.K and Roy J (2004). Antimicrobial and chemopreventive properties of herbs and spices. Curr. Med. Chem, 11 (11): 1451–1460.
Livermore D.M (2004). The need for new antibiotics. Clinical Microbiology & Infection, 4(10): 1-9.
Mascaretti O.A (2003). Bacteria versus antibacterial agents: An integrated approach. Washington: ASM Press.
Nally J.D (Ed.) (2007). Good manufacturing practices for pharmaceuticals. Sixth edition. Informa Healthcare USA, Inc, New York.
Discover more from Microbiology Class
Subscribe to get the latest posts sent to your email.