The polymerase chain reaction (PCR) is one of the most powerful technologies in molecular biology. Using PCR, specific sequences within a DNA or complementary DNA (cDNA) template can be copied, or “amplified”, many thousand- to a million-fold using sequence-specific oligonucleotides (or primers), heat-stable DNA polymerase, and thermal cycling. In traditional (endpoint) PCR, detection and quantification of the amplified sequence are performed at the end of the reaction after the last PCR cycle, and involve post-PCR analysis such as gel electrophoresis and image analysis. In real-time quantitative PCR (qPCR), PCR product is measured at each cycle. By monitoring reactions during the exponential amplification phase of the reaction, users can determine the initial quantity of the target with great precision.
PCR theoretically amplifies DNA exponentially, doubling the number of target molecules with each amplification cycle. When it was first developed, scientists reasoned that the number of cycles and the amount of PCR end-product could be used to calculate the initial quantity of genetic material by comparison with a known standard. To address the need for robust quantification, the technique of real-time quantitative PCR was developed. Currently, endpoint PCR is used mostly to amplify specific DNA for sequencing, cloning, and use in other molecular biology techniques.
In real-time PCR, the amount of DNA is measured after each cycle via fluorescent dyes that yield increasing fluorescent signal in direct proportion to the number of PCR product molecules (amplicons) generated. Data collected in the exponential phase of the reaction yield quantitative information on the starting quantity of the amplification target. Fluorescent reporters used in real-time PCR include double-stranded DNA (dsDNA)–binding dyes, or dye molecules attached to PCR primers or probes that hybridize with PCR products during amplification.
The change in fluorescence over the course of the reaction is measured by an instrument that combines thermal cycling with fluorescent dye scanning capability. By plotting fluorescence against the cycle number, the real-time PCR instrument generates an amplification plot that represents the accumulation of product over the duration of the entire PCR reaction (Figure 1).
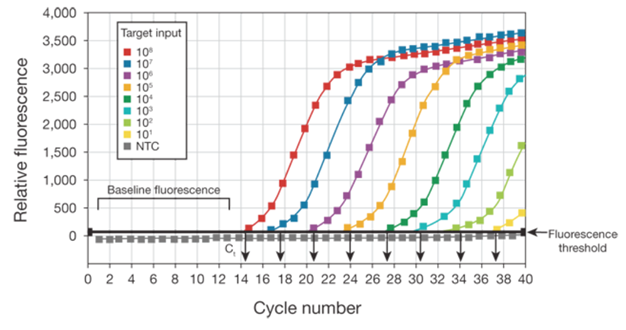
As shown in Figure 1, amplification plots are created when the fluorescent signal from each sample is plotted against
cycle number; therefore, amplification plots represent the accumulation of product over the duration of the real-time PCR experiment. The samples used to create the plots in this figure are a dilution series of the target DNA sequence.
The advantages of real-time PCR include:
- Ability to monitor the progress of the PCR reaction as it occurs in real time.
- Ability to precisely measure the amount of amplicon at each cycle, which allows highly accurate quantification of the amount of starting material in samples.
- An increased dynamic range of detection.
- Amplification and detection occur in a single tube, eliminating post-PCR manipulations.
Over the past several years, real-time PCR has become the leading tool for the detection and quantification of DNA or RNA. Using these techniques, you can achieve precise detection that is accurate within a 2-fold range, with a dynamic range of input material covering 6 to 8 orders of magnitude.
Source
www.lifetechnologies.com
Discover more from #1 Microbiology Resource Hub
Subscribe to get the latest posts to your email.


