Real-time PCR fluorescence detection systems
Real-time fluorescent PCR chemistries
Many real-time fluorescent PCR chemistries exist, but the most widely used are 5” nuclease assays such as TaqMan®
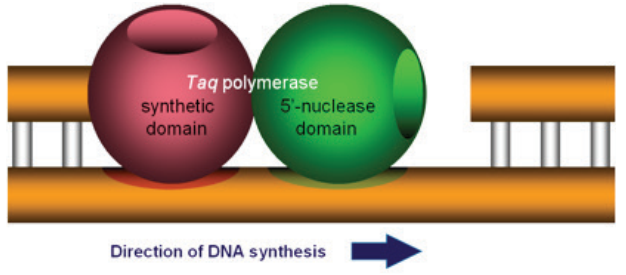
Assays and SYBR® Green dye–based assays (Figure 1). The 5” nuclease assay is named for the 5” nuclease activity
associated with Taq DNA polymerase (Figure 2). The 5” nuclease domain has the ability to degrade DNA
bound to the template, downstream of DNA synthesis.
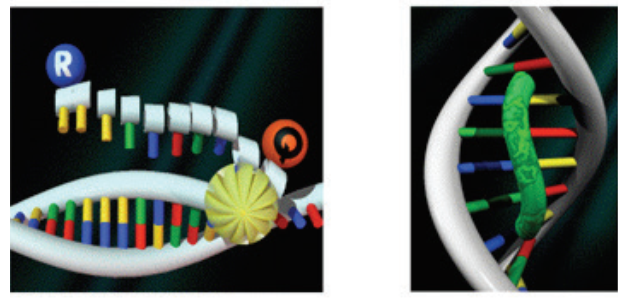
A second key element in the 5” nuclease assay is a phenomenon called fluorescence resonance energy transfer (FRET). In FRET, the emissions of a fluorescent dye can be strongly reduced by the presence of another dye, often called the quencher, in close proximity. FRET can be illustrated by two fluorescent dyes: green and red (Figure 3). The green fluorescent dye has a higher energy of emission compared to the red, because green light has a shorter wavelength compared to red.
dye binding to DNA (right)
If the red dye is in close proximity to the green dye, excitation of the green dye will cause the green emission energy to be transferred to the red dye. In other words, energy is being transferred from a higher to a lower level. Consequently, the signal from the green dye will be suppressed or “quenched”. However, if the two dyes are not in close proximity, FRET cannot occur, allowing the green dye to emit its full signal.
sphere represents a protein domain.
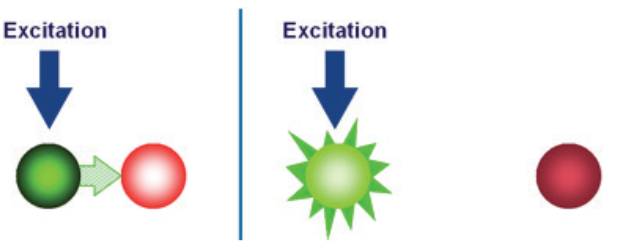
a green light–emitting fluorescent dye is in close proximity to a red
light–emitting fluorescent dye. (B) FRET does not occur when the two
fluorescent dyes are not in close proximity.
A 5” nuclease assay for target detection or quantification typically consists of two PCR primers and a TaqMan® probe (Figure 4). Before PCR begins, the TaqMan® probe is intact and has a degree of flexibility. While the probe is intact, the reporter and quencher have a natural affinity for each other, allowing FRET to occur (Figure 5). The reporter signal is
quenched prior to PCR. During PCR, the primers and probe anneal to the target.
DNA polymerase extends the primer upstream of the probe. If the probe is bound to the correct target sequence, the polymerase’s 5” nuclease activity cleaves the probe, releasing a fragment containing the reporter dye. Once cleavage takes place, the reporter and quencher dyes are no longer attracted to each other; the released reporter molecule will no longer be quenched.
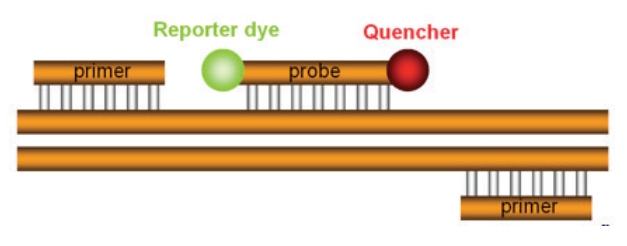
sequence and is designed to bind the target between the two PCR
primers. Attached to the 5” end of the TaqMan® probe is the “reporter”,
which is a fluorescent dye that will report the amplification of the
target. On the 3” end of the probe is a quencher, which quenches fluorescence from the reporter in intact probes. The quencher also blocks
the 3” end of the probe so that it cannot be extended by thermostable
DNA polymerase.
5” nuclease assay specificity
Assay specificity is the degree that the assay includes signal from the target and excludes signal from non-target in the results. Specificity is arguably the most important aspect of any assay. The greatest threat to assay specificity for 5” nuclease assays is homologs. Homologs are genes similar in sequence to that of the target, but they are not the intended target of the assay. Homologs are extremely common within species and across related species.
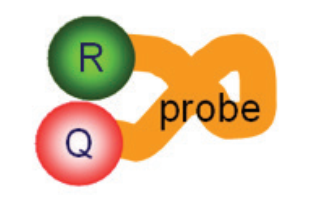
reporter dye, Q is the quencher molecule, and the orange line
represents the oligonucleotide.
5” nuclease assays offer two tools for specificity: primers and probes. A mismatch between the target and homolog positioned at the 3”-most base of the primer has maximal impact on the specificity of the primer. A mismatch further away from the 3” end will have less impact on specificity. In contrast, mismatches across most of the length of a TaqMan® MGB probe, which is shorter than a TaqMan® TAMRA™ probe, can have a strong impact on specificity—TaqMan® MGB probes are stronger tools for specificity than primers.
For example, a 1- or 2-base random mismatch in the primer binding site will very likely allow the DNA polymerase to extend the primer bound to the homolog with high efficiency. A one or two base extension by DNA polymerase will stabilize the primer bound to the homolog, so it is just as stably bound as primer bound to the intended, fully complementary target. At that point, there is nothing to prevent the DNA polymerase from continuing synthesis to produce a copy of the homolog.
In contrast, mismatches on the 5” end of the TaqMan® probe binding site cannot be stabilized by the DNA polymerase due to the quencher block on the 3” end. Mismatches in a TaqMan® MGB probe binding site will reduce how tightly the probe is bound, so that instead of cleavage, the intact probe is displaced. The intact probe returns to its quenched configuration, so that when data are collected at the end of the PCR cycle, signal is produced from the target but not from the homolog, even though the homolog is being amplified. In addition to homologs, PCR may also amplify nonspecific products produced by primers binding to seemingly random locations in the sample DNA or sometimes to themselves in so-called “primer-dimers”. Since nonspecific products are unrelated to the target, they do not have TaqMan® probe binding sites, and thus are not seen in the real-time PCR data.
TaqMan® probe types
TaqMan® probes may be divided into two types: MGB and non-MGB. The first TaqMan® probes could be classified as non-MGB. They used a dye called TAMRA™ dye as the quencher. Early in the development of real-time PCR, extensive testing revealed that TaqMan® probes required an annealing temperature significantly higher than that of PCR primers to allow cleavage to take place. TaqMan® probes were therefore longer than primers. A 1-base mismatch in such long probes had a relatively mild effect on probe binding, allowing cleavage to take place. However, for many applications involving high genetic complexity, such as eukaryotic gene expression and single nucleotide polymorphisms (SNPs), a higher degree of specificity was desirable.
TaqMan® MGB probes were a later refinement of the TaqMan® probe technology. TaqMan® MGB probes possess a minor-groove binding (MGB) molecule on the 3” end. Where the probe binds to the target, a short minor groove is formed in the DNA, allowing the MGB molecule to bind and increase the melting temperature, thus strengthening probe binding.
Consequently, TaqMan® MGB probes can be much shorter than PCR primers. Because of the MGB moiety, these probes can be shorter than TaqMan® probes and still achieve a high melting temperature. This enables TaqMan® MGB probes to bind to the target more specifically than primers at higher temperatures. With the shorter probe size, a 1-base mismatch has a much greater impact on TaqMan® MGB probe binding. And because of this higher level of specificity, TaqMan® MGB probes are recommended for most applications involving
high genetic complexity.
Source
www.lifetechnologies.com
Discover more from Microbiology Class
Subscribe to get the latest posts sent to your email.