Based on their effects on the structural integrity of the DNA molecule, mutations can be classified as substitution, insertion, deletion, inversion, reciprocal translocation and chromosomal rearrangements.
SUBSTITUTION (BASE-PAIR SUBSTITUTION)
Substitution literally means the act of replacing one thing with another. When base substitution as a type of mutation occurs during DNA replication, a single base at one point in the DNA replication process is replaced by one of the other three bases. This type of mutation can also be called point mutation. And there are various types of base substitution including silent mutation, missense mutation, and frameshift mutation. Base substitution during DNA replication causes silent, missense, nonsense mutation and frameshift mutational effects (Figure 1). It is also noteworthy that the consequences of base-pair substitution mutations in protein coding regions of a DNA or gene depend on the type of substitution and the location where it occurred.
Silent mutation is a type of mutation that does not affect the phenotype of the cell undergoing it. It usually occurs outside of the gene. Though they occur outside of a gene and may not produce a mutant phenotype, silent mutations can also be observed within the gene of a cell. Silent mutation also occurs within the coding region of a gene as aforementioned. For example, arginine (ARG) is an amino acid with two different codons viz: AGA and AGG; and thus a substitution in an AGA codon to AGG would have no effect on the amino acid – since both codons (AGA and AGG) code for the same amino acid (which is arginine). In general, a base-pair substitution can occur either inside the gene or outside the gene.
And a base substitution that occurs within the protein-coding region of a gene will often result in the miscoding of an amino acid residue; and this will result in the synthesis of a mutant protein molecule. Such mutant protein molecules may have a partial loss of its normal biological function and or an occasional increase in its biological activity. If a point mutation (base-pair substitution) occurs within the coding region of a gene that encodes a particular polypeptide, any change can result in a change in the amino acid sequence of the polypeptide molecule, and such mutational changes has little or no effect on the phenotype of the cell since not all mutations in the base sequence encoding a polypeptide will change the polypeptide.
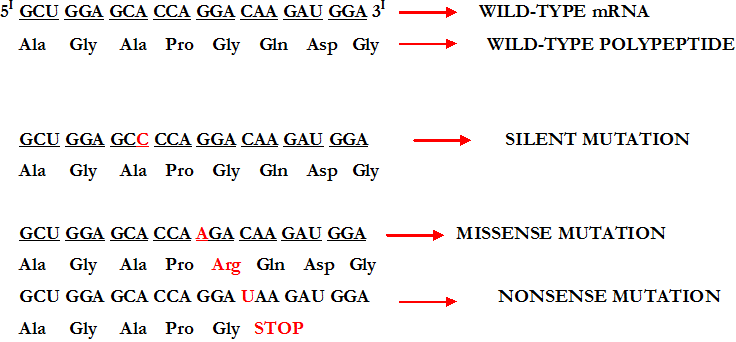
Key: Ala=Alanine, Gly=Glycine, Pro=Proline, Asp=Aspartic acid, Arg=Arginine
Silent mutation does not change an amino acid. But in some scenarios, silent mutations can still produce a phenotypic effect on the protein synthesis process either by speeding up or slowing down protein synthesis or by effecting gene splicing. Missense mutation is a mutation in which a base substitution could result in an amino acid substitution. Unlike in silent mutations in which there is no novel amino acid in the protein sequence as a result of the genetic modification, the production of a new amino acid molecule usually accompanies missense mutation.
Missense mutation has tremendous biological consequences. It is a single base change that results in the change of an amino acid within a given polypeptide molecule. For example, the codon CTC in the DNA sense strand (and GAG in mRNA) specifies a glutamate residue in the protein molecule. But when this codon (i.e. CTC or GAG) is altered in the DNA sense strand and mRNA strand to CAC and GUG respectively, a different amino acid molecule (in this case valine) is produced. In missense mutations, there is usually the addition or insertion of the wrong amino acid molecule.
Sickle cell anaemia is a typical example of a genetic disease or disorder due to missense mutation. Sickle cell disease is a blood borne genetic/hereditary disease in which the sufferers have a dysfunctional type of red blood cells that resembles a sickle. Missense mutation changes an amino acid to a different amino acid molecule. Depending on the biological function of the affected amino acid molecule, missense mutation may affect the protein function which those affected amino molecules encodes.
Nonsense mutation is a type of mutation in which a prematurely type of shortened protein molecule is formed. In nonsense mutation, the protein formed is usually truncated and an incomplete protein molecule is formed. Nonsense mutations generally result in the production of a STOP CODON; and it changes an amino acid to a STOP CODON so that the protein synthesis process will automatically stop. Nonsense mutation as shown in Figure 1 result in the premature termination of translation.
Mutations in which STOP CODONS are formed are generally known as nonsense mutations. Nonsense mutation creates a STOP CODON in the middle of a gene, and this leads to the formation of an incomplete protein molecule. Frameshift mutation is a type of mutation that occurs within the protein coding region of a gene. And such mutagenesis or mutations usually arise from the addition or deletion of one or few bases that are not multiples of three. Frameshift mutations include deletions, insertion and gene duplication that usually occur during DNA replication.
Codons are normally in the groups of three nucleotides (i.e. codons are made of three nucleotide bases), and when this pattern is not followed, there will be an alteration in the reading frame of the gene. This type of mutation that occurs in the translational reading frame of a gene is known as a frameshift mutation. There are only three possible reading frames for each gene since codons are groups of three nucleotides. A codon is a group of three successive nucleotides found in the mRNA; and they base-pair with the anticodon of an individual tRNA that carries a specific amino acid molecule.
A deletion or insertion (i.e. frameshift mutation) of a number of bases that is not a multiple of three usually introduces premature STOP codons that inhibit the protein synthesis process in addition to other genetic alterations associated with frameshift mutations. Reading frames other than the correct translational reading frames often contain STOP codons which will truncate the mutant protein prematurely. Frameshift mutations completely render mutant protein molecules nonfunctional.
The removal of a base (as underlined in Figure 1) results in a change in the coding of the adjacent bases sequences, and this produces a highly altered (mutant) protein molecule (Figure 2). Frameshift mutations almost always cause long stretches of altered amino acids resulting in the production of inactive protein molecules as aforementioned; and this type of mutation is generally known to delete or insert one or a few nucleotide bases in the translational reading frame of an mRNA molecule.
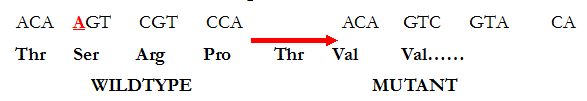
Key: Thr=Threonine, Ser=Serine, Arg=Arginine, Pro=Proline, Val=Valine
A reading frame refers to one of three possible ways of reading a nucleotide sequence. It is a way of dividing the sequence of nucleotides in a nucleic acid molecule (DNA or RNA) molecule into a set of consecutive, non-overlapping triplets general known as codons. There are three reading frames that can be read in the 5’→3′ direction of DNA synthesis and each of these begins from a different nucleotide in a triplet known as a codon. The reading frame affects which protein is made during translation.
If for example we have a 15 DNA base pairs as illustrated below:
ACTTAGCCGGGACTA
- We can start reading or translating the DNA from the first letter, ‘A,’. This first alphabet would be referred to as the first reading frame of the translation process.
- We can also start to translate the DNA from the second letter, ‘C,’; and this would be referred to as the second reading frame.
- And we can also start reading or translating from the third letter, ‘T,’; and this would be referred to as the third reading frame.
In all, there are actually six reading frames. Three of the reading frames are on the positive strand of the DNA while three of the reading frame (which is read in the reverse direction) is found on the negative strand. It is noteworthy that the first three reading frames as aforementioned are read in the forward direction.
INSERTION
Insertion is a type of mutation that occurs when there is a gain of nucleotide base pairs. It occurs when there is an addition of one or more nucleotide bases into a DNA sequence. When insertions occur in the amino acid coding region of a gene (i.e. the exon), it can cause genetic alterations that will hazardously affect the resulting phenotype. Insertions and the subsequent frameshift mutation that occur in the reading frame will cause the active translation of the gene to encounter a premature STOP codon, and this will result in an end to translation and the production of a truncated protein molecule.
DELETION
Deletion is a type of mutation that results in the loss of nucleotide base pairs. Deletion mutation is a type of mutagenesis in which a part of a chromosome or a sequence of DNA is lost during DNA replication. Deletions that do not occur in multiples of three bases can cause a frameshift mutation by changing the 3-nucleotide protein reading frame of the genetic sequence.
Deletion of a number of pairs that is not evenly divisible by three will lead to a frameshift mutation, causing all of the codons occurring after the deletion to be read incorrectly during translation, producing a severely altered and potentially nonfunctional protein molecule. Small deletions are usually less likely to be fatal but large deletions are usually fatal, and may cause several genetic disorders in the host. Deletion mutation removes the segment of a DNA molecule; and this development can result in the loss of substantial segments of the chromosome.
INVERSION
Inversion mutation is a type of mutation or gene rearrangement that occur when the normal order of a gene sequence is flipped in such a way that the chromosomal segment are placed in the opposite orientation with respect to other chromosomes. Inversions are chromosome rearrangements in which a segment of a DNA molecule is reversed end to end. For example, a given chromosome segment depicted as: “abcdefgh” can be inverted or rearranged to be: “ab-edc-fg-h”. Inversions usually do not cause any abnormalities in carriers as long as the rearrangement is balanced with no extra or missing DNA.
CHROMOSOMAL REARRANGEMENTS
Chromosomal rearrangements involve gene deletions, inversions, duplications and translocations. And they usually arise from breakages that occur in the DNA. When there is a break in the DNA structure, a rejoining of the broken DNA can result in the production of a novel chromosomal arrangement of genes that are quite different from the normal order of the gene before the breakage occurred.
Studying mutations in living organisms inclusive of microorganisms is important because changes that affect the entire chromosome or some segments of the chromosomes can cause significant problems associated with the organism’s growth, development and other body functions. Our understanding of mutations in living systems and how cells can be genetically manipulated and/or mutated or transformed will help us to understand more complex biological processes that occur in living cells such as carcinogenesis.
Mutations can also be classified based on their effect on the encoded protein since mutations outside the coding sequence of the gene can also impact on the outcome of gene expression. The DNA or gene encodes the genetic information for the production of a particular protein molecule in the cell of a living organism. Any alteration in the nucleotide base sequence(s) of the gene will ultimately affect the outcome of the protein to be synthesized by the cell. There could be complete loss of function or gain of function when the mutation impact on the protein functions.
References
Bains W (1998). Biotechnology: From A to Z. 2nd ed. Oxford University Press, New York, USA.
Bourgaize D, Jewell T.R and Buiser R.G (1999). Biotechnology: Demystifying the Concepts. Pearson Education, San Francisco, CA.
Brian Robert Shmaefsky (2006). Biotechnology 101. Greenwood Publishing Group, Inc, USA. Pp. 1-273.
Bushell M.E (1998). Application of the principles of industrial microbiology to biotechnology (ed. Wiseman, A.) Chapman and Hall, New York. Pp. 5–43.
Byong H. Lee (2015). Fundamentals of Food Biotechnology. Second edition. Wiley-Blackwell, New Jersey, United States.
Chrispeels M.J and Sadava D.E (2002). Plants, Genes, and Crop Biotechnology. 2nd edition. Jones and Bartlett Publishers, Sudbury, MA.
Clark D.P and Pazdernik N (2010). Biotechnology. First edition. Elsevier Science and Technology Books, Amsterdam, Netherlands.
Das H.K (2010). Textbook of Biotechnology. Fourth edition. Wiley edition. Wiley India Pvt, Ltd, New Delhi, India.
Dictionary of Microbiology and Molecular Biology, 3rd Edition. Paul Singleton and Diana Sainsbury. 2006, John Wiley & Sons Ltd. Canada.
Glick B.R and Pasternak J.J (2003). Molecular Biotechnology: Principles and Applications of Recombinant DNA. ASM Press, Washington DC, USA.
Godbey W.T (2014). An Introduction to Biotechnology. First edition. Woodhead Publishing, Cambridge, United Kingdom.
Jee C and Shagufta (2007). Environmental Biotechnology. APH Publishing Corporation, Darya Ganj, New Delhi, India.
Lee S.Y, Lee D.Y and Kim T.Y (2005). Systems biotechnology for strain improvement. TRENDS in Biotechnology, 23(7):349-356.
Discover more from Microbiology Class
Subscribe to get the latest posts sent to your email.